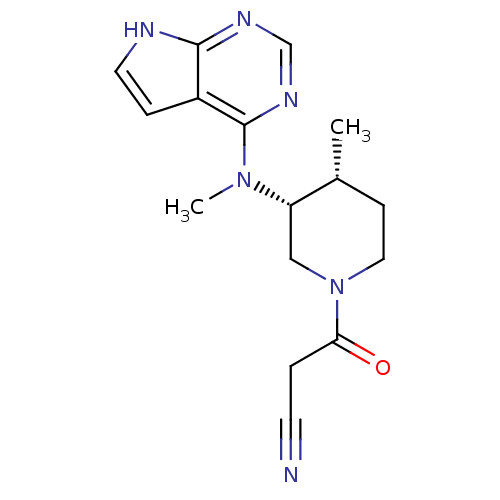
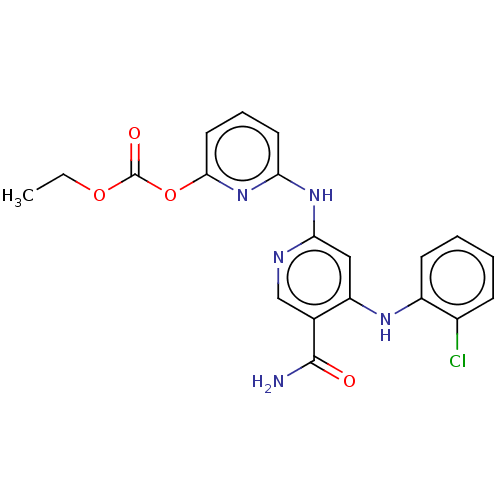
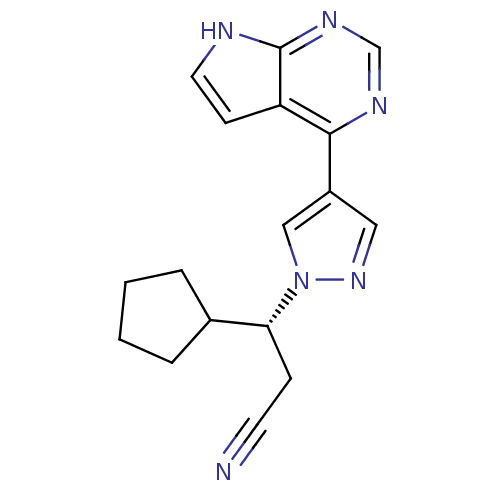
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
Target InfoPDBNCI pathwayReactome pathwayKEGG
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
In DepthDetails
Target InfoPDBMMDBReactome pathwayKEGG
UniProtKB/SwissProtUniProtKB/TrEMBL
B.MOADDrugBankantibodypediaGoogleScholar
UniProtKB/SwissProtUniProtKB/TrEMBL
B.MOADDrugBankantibodypediaGoogleScholar
In DepthDetails
Target InfoPDBMMDBNCI pathwayReactome pathwayKEGG
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
In DepthDetails
Target InfoPDBMMDBNCI pathwayReactome pathwayKEGG
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
In DepthDetails
Target InfoPDBMMDBNCI pathwayReactome pathwayKEGG
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
UniProtKB/SwissProt
B.MOADDrugBankantibodypediaGoogleScholar
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails
In DepthDetails