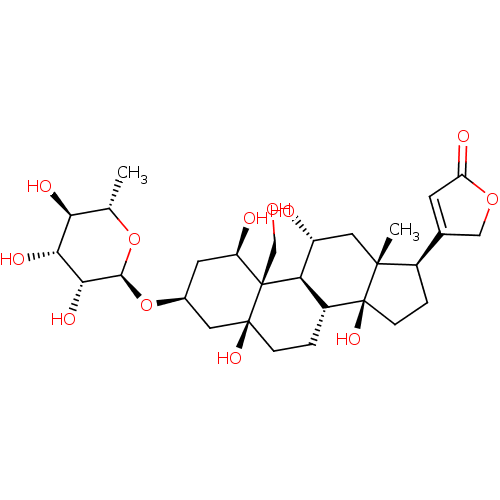
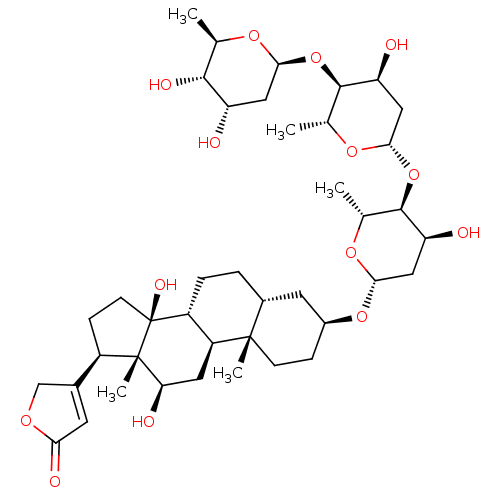
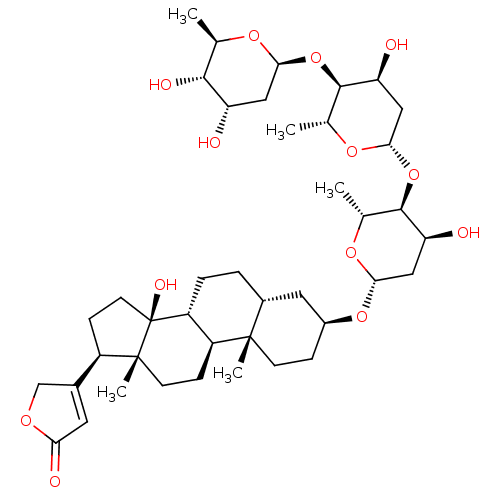
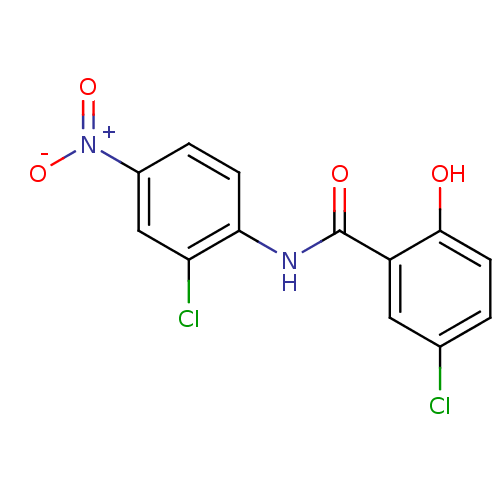
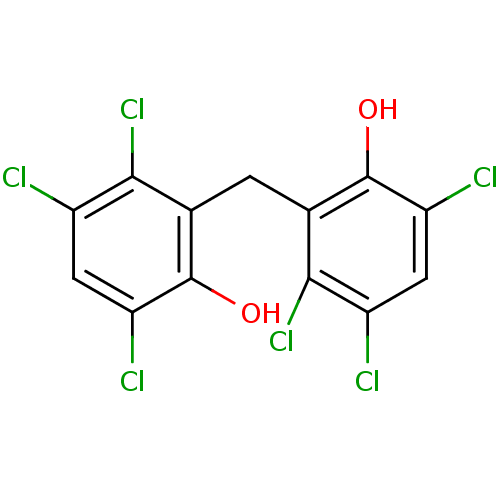
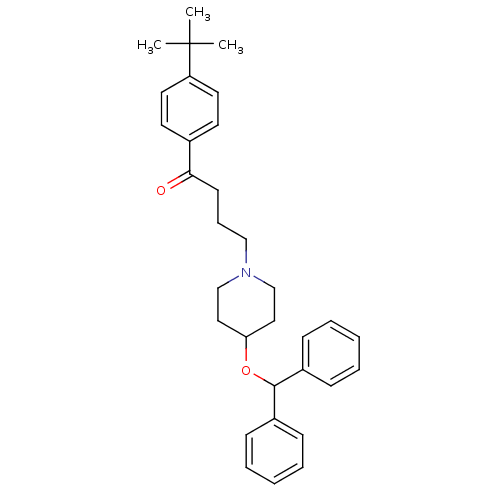
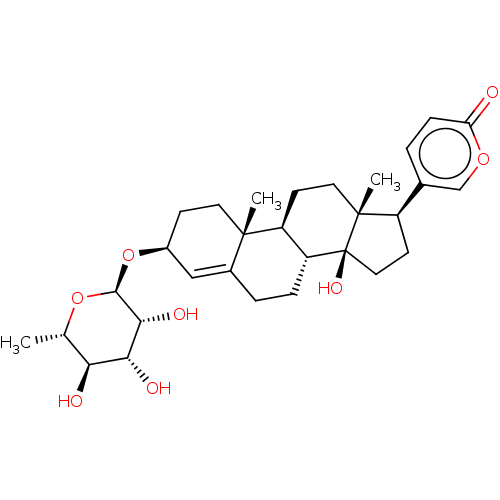
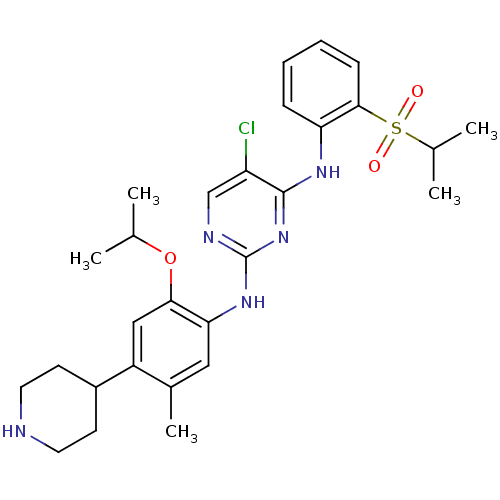
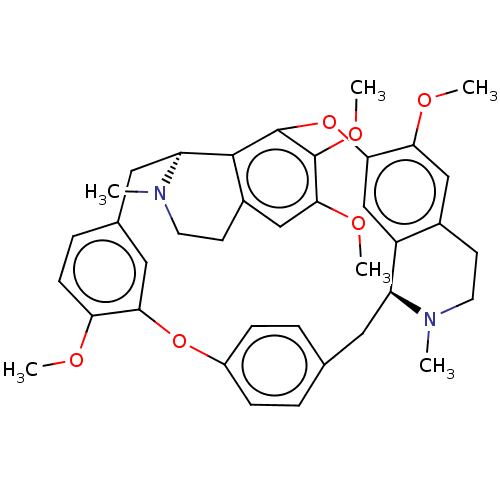
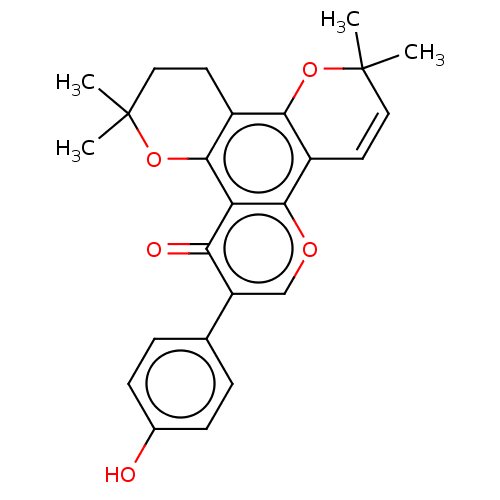
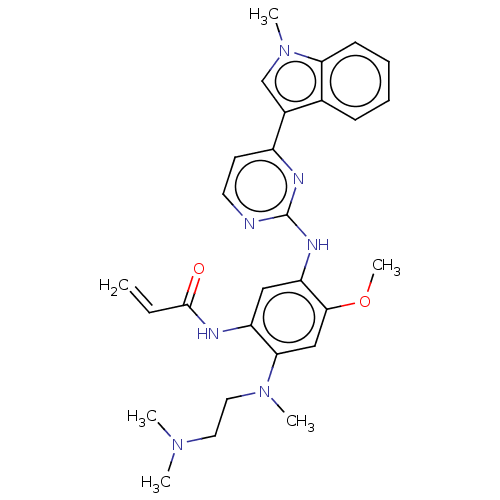
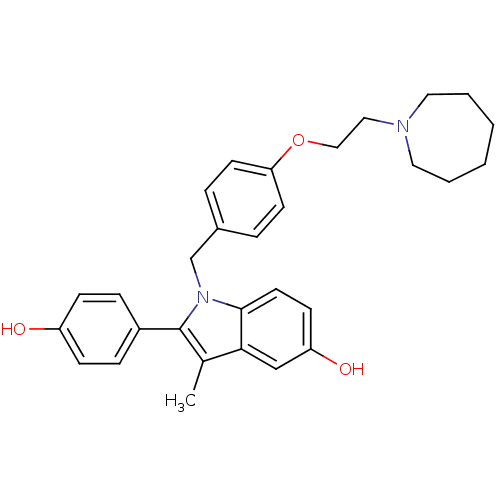
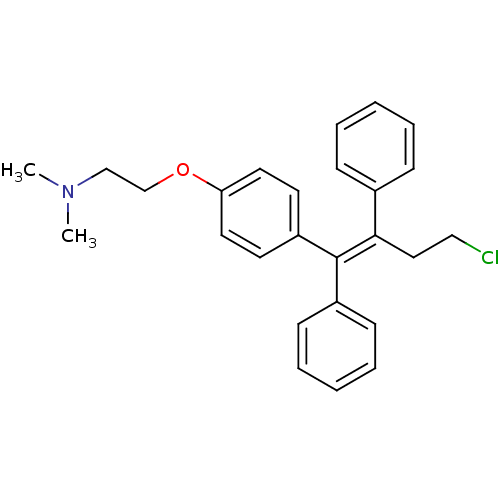
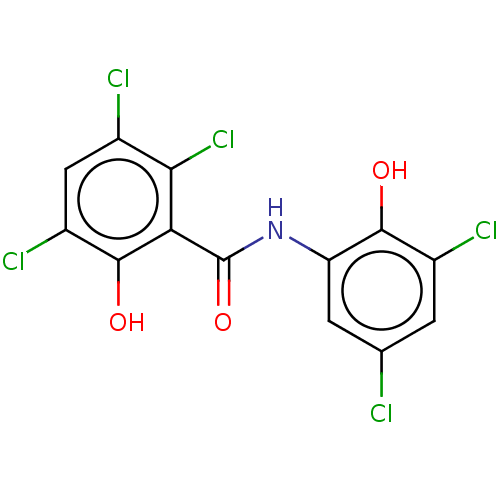
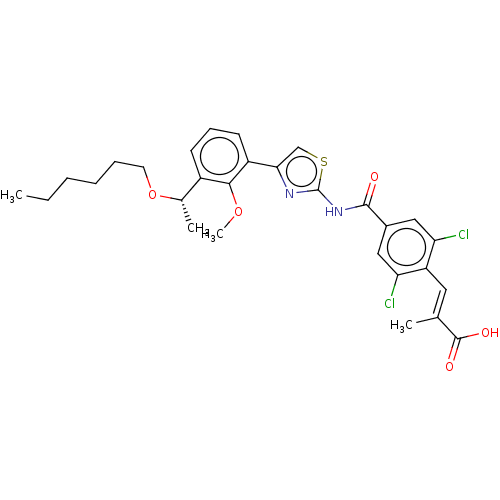
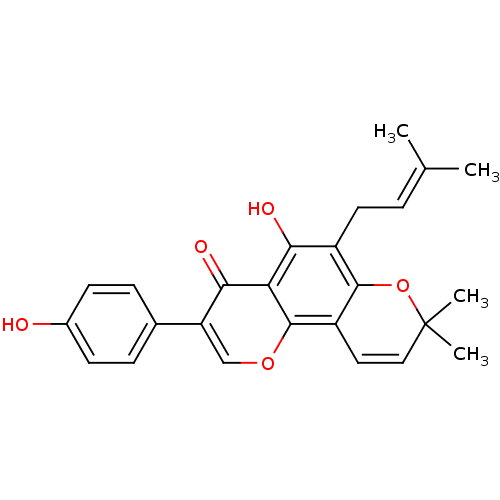
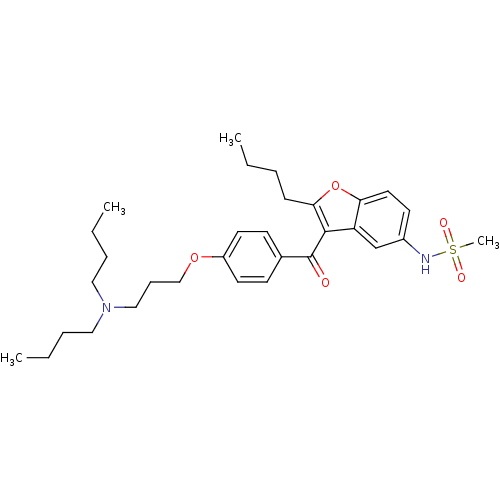
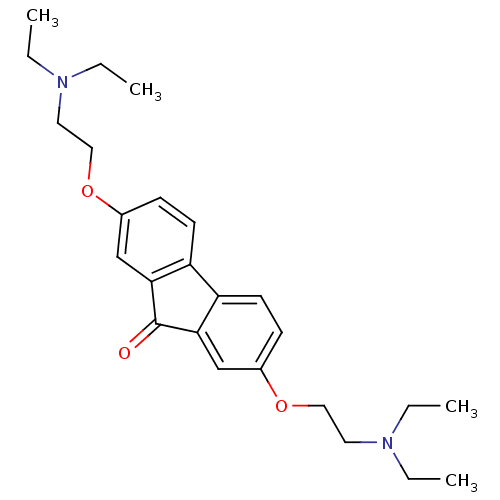
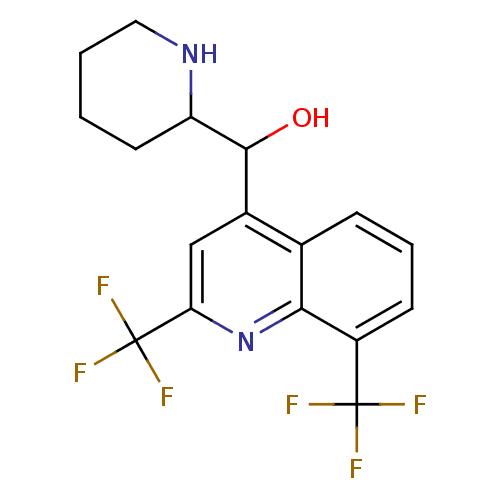
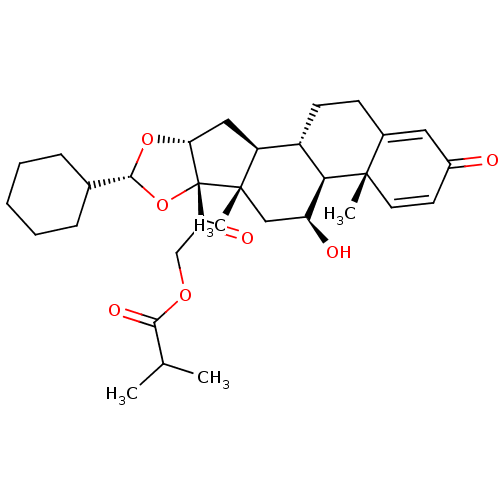
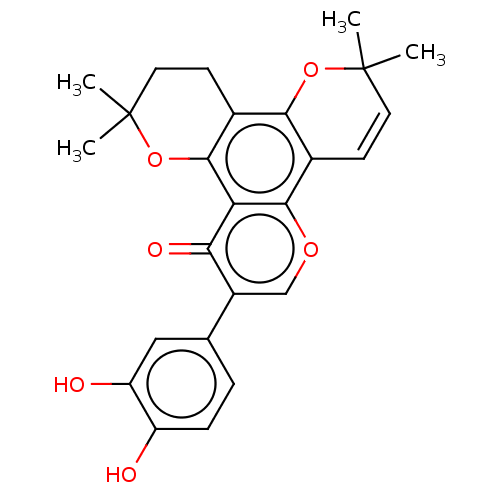
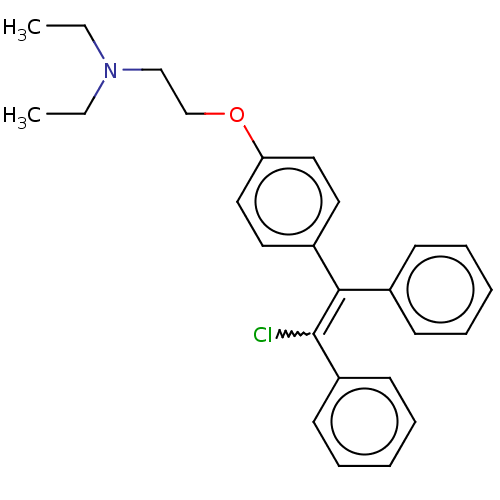
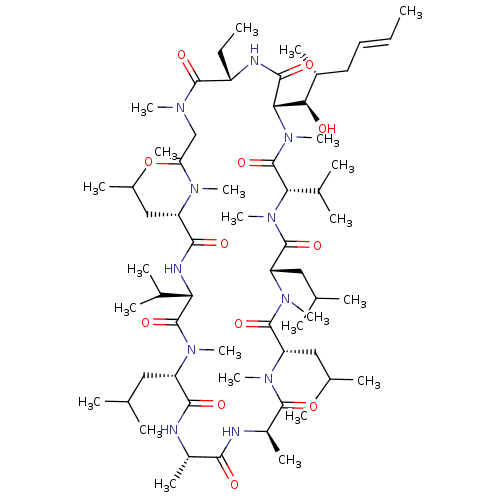
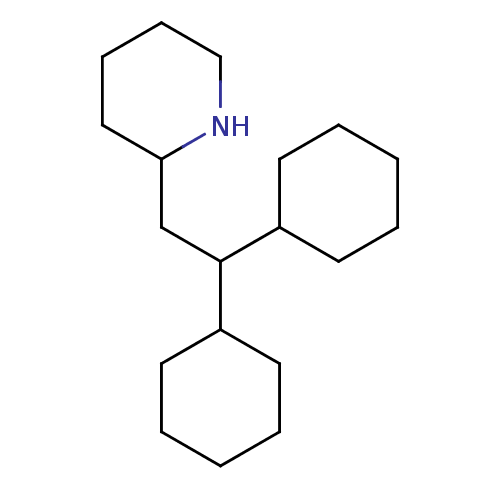
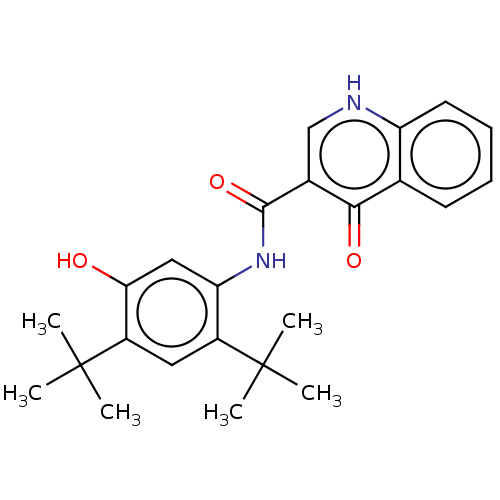
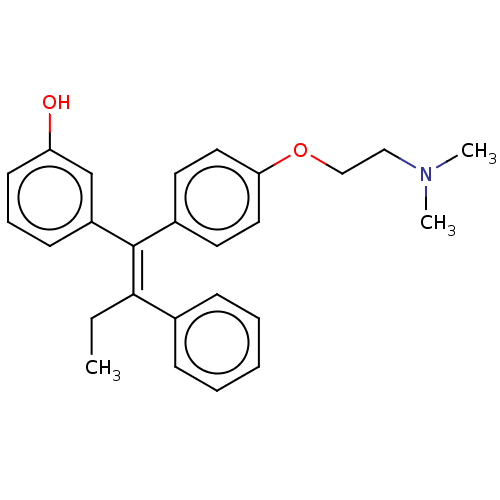
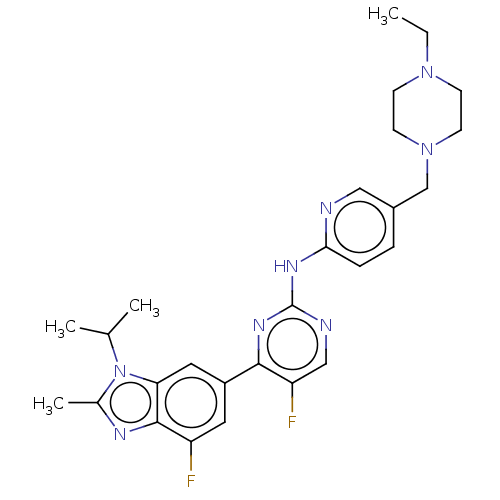
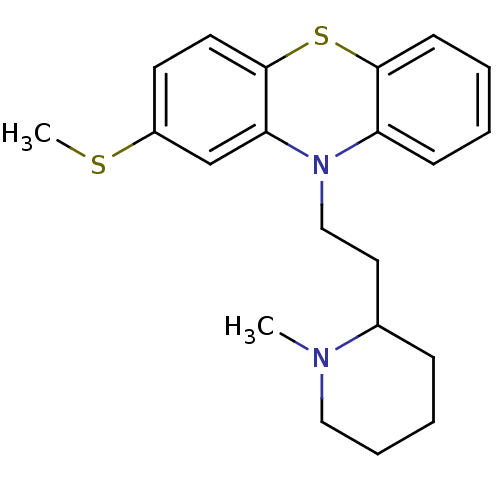
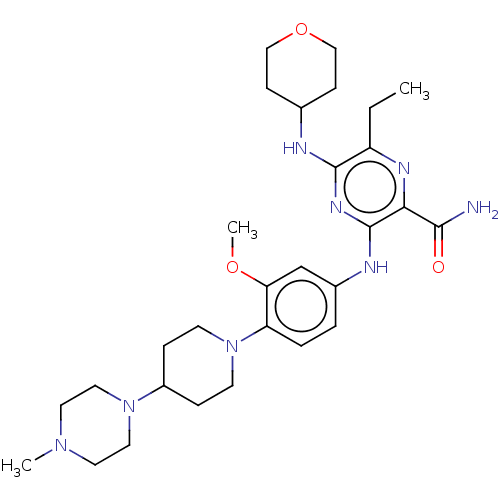
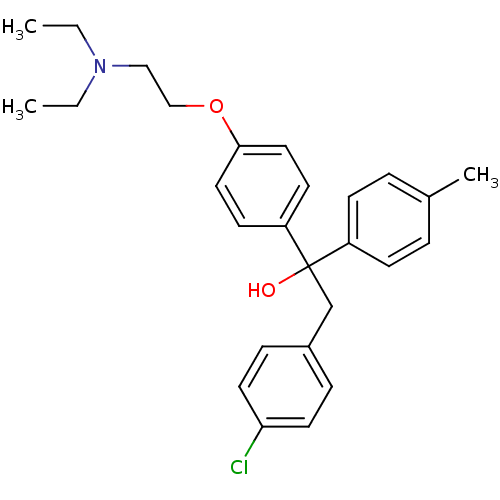
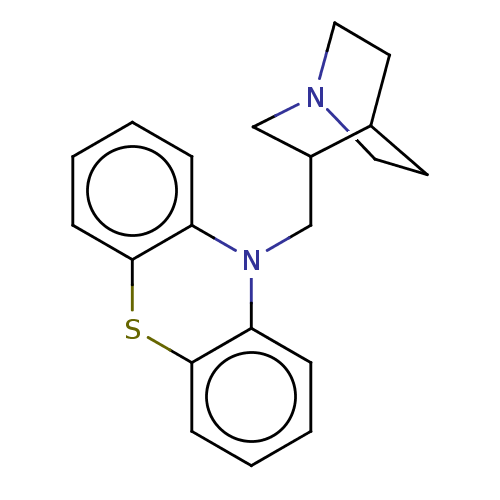
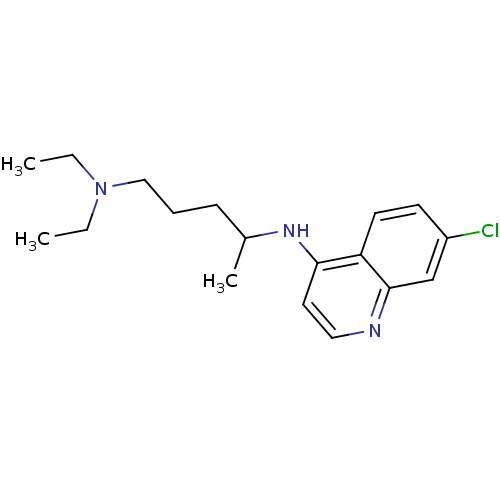
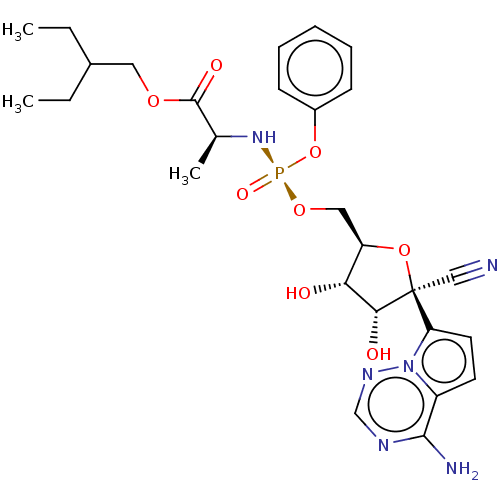
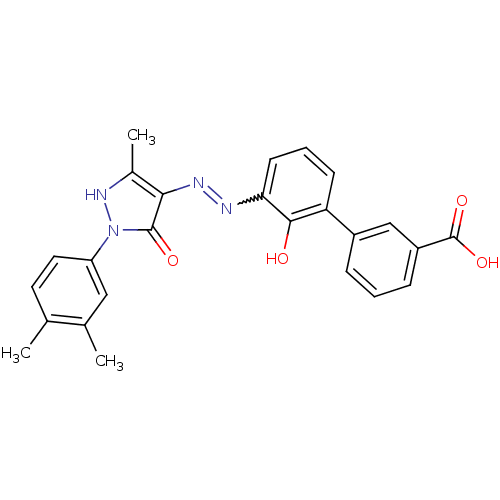
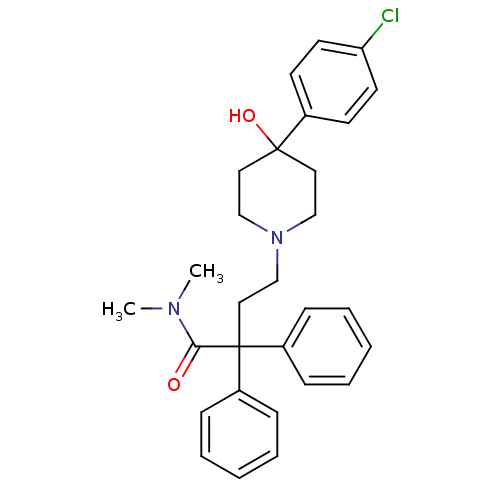
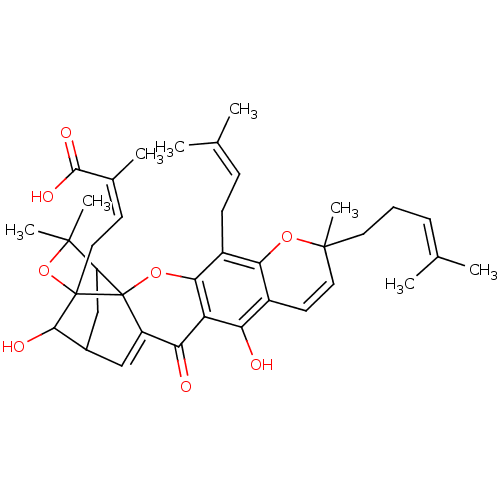
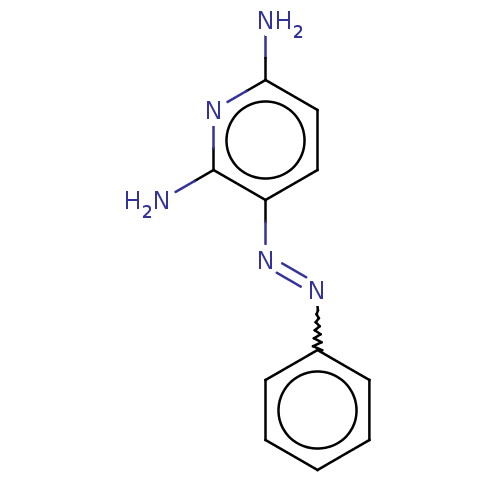
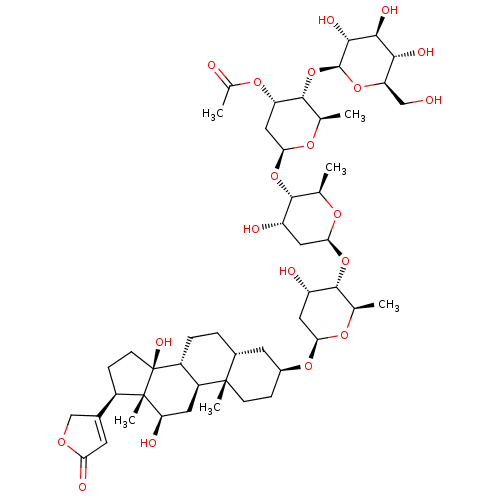
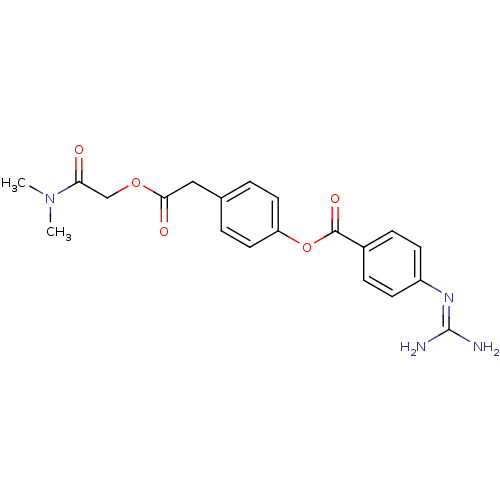
Affinity DataIC50: 97nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 190nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 230nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 240nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 280nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 900nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 920nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.04E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.86E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.22E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.26E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.44E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.58E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.71E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.78E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.87E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.92E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.09E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.33E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.33E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.47E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.51E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.64E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.01E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.15E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.36E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.82E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.30E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.38E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.57E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.60E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.62E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.69E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.76E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.05E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.28E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.28E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.87E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 8.24E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 8.27E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 9.12E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 9.27E+3nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.14E+4nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+4nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.34E+4nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.53E+4nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.80E+4nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+4nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+4nMAssay Description:Ten-point DRCs were generated for each drug. Vero cells were seeded at 1.2 × 104 cells per well in DMEM, supplemented with 2% FBS and 1× ...More data for this Ligand-Target Pair