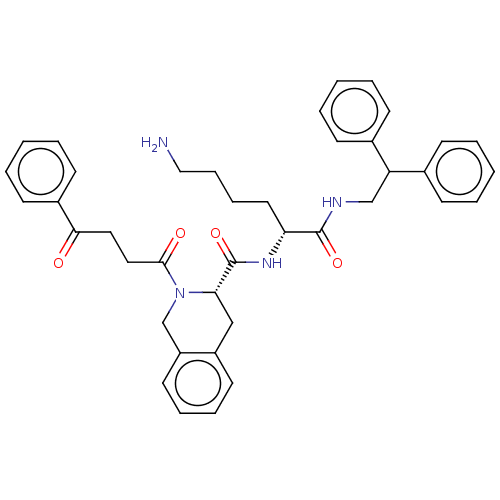
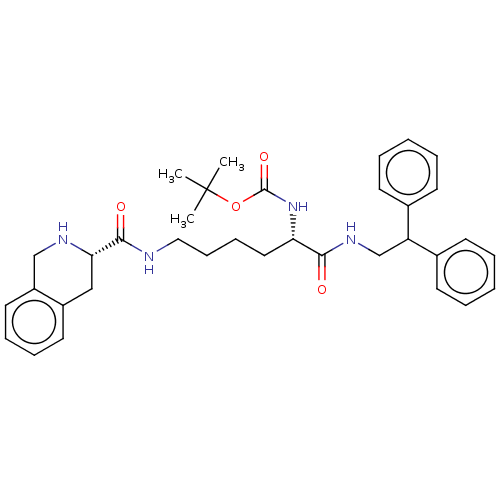
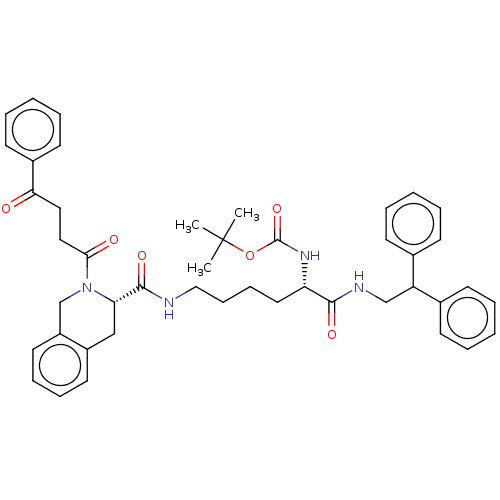
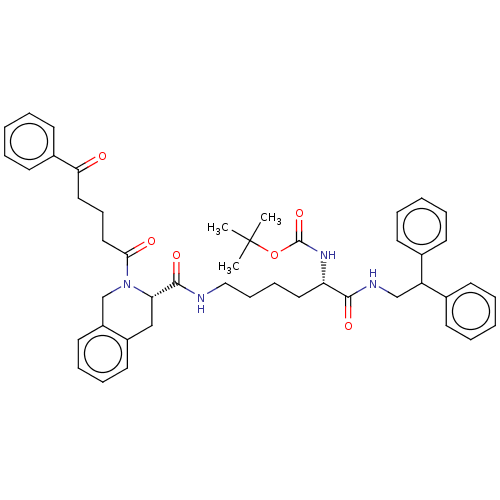
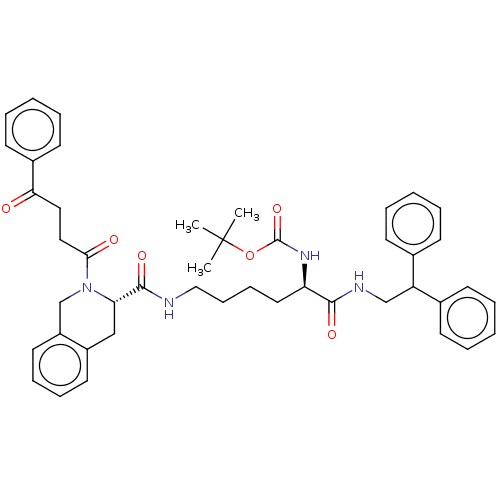
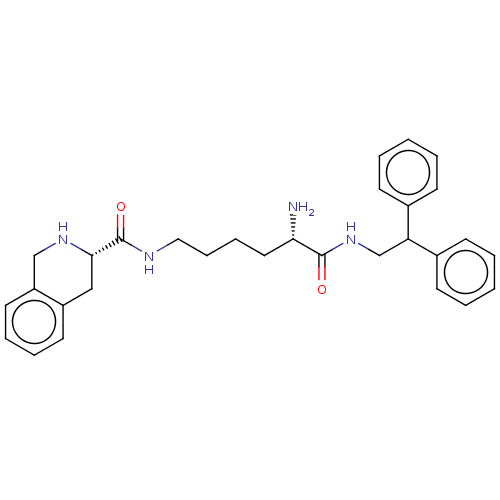
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 59nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 759nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 194nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 263nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 229nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 195nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 126nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 166nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 195nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 302nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 138nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 282nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair
TargetC-X-C chemokine receptor type 3(Homo sapiens (Human))
Friedrich Alexander University
Curated by ChEMBL
Friedrich Alexander University
Curated by ChEMBL
Affinity DataEC50: 309nMAssay Description:Positive allosteric modulation of ProLink2-tagged wild type CXCR3 (unknown origin) expressed in HEK293T cells assessed as beta-arrestin-2 recruitment...More data for this Ligand-Target Pair