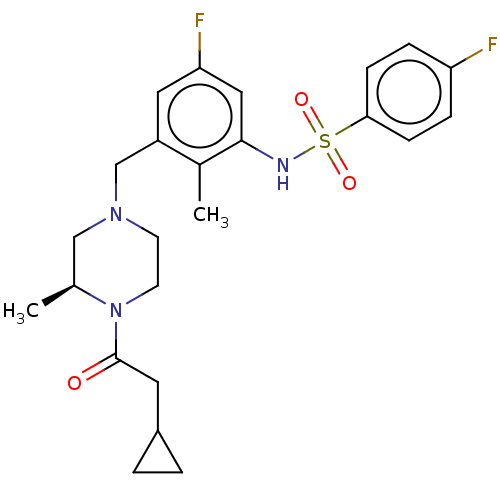
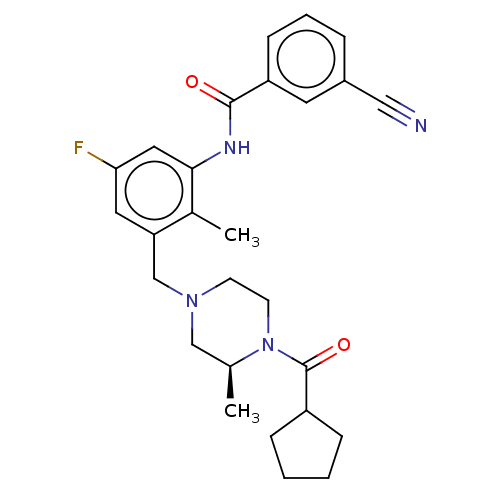
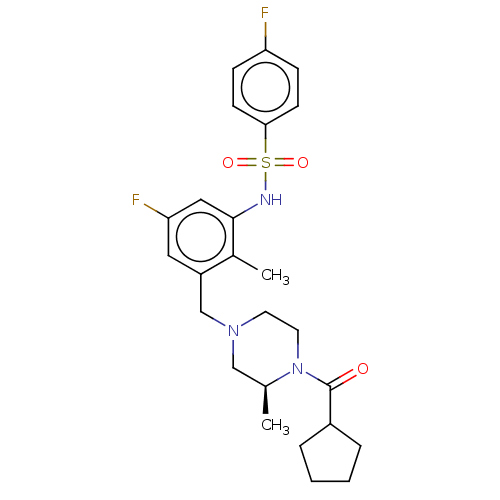
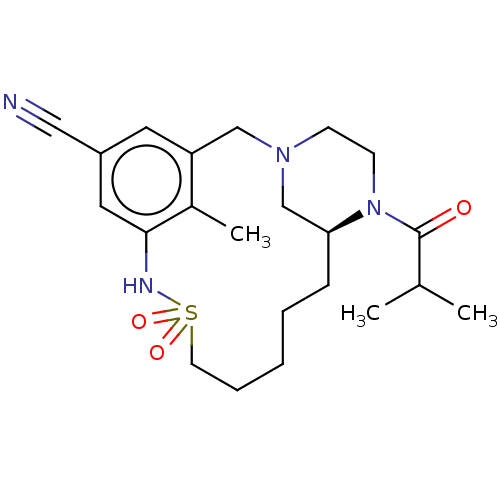
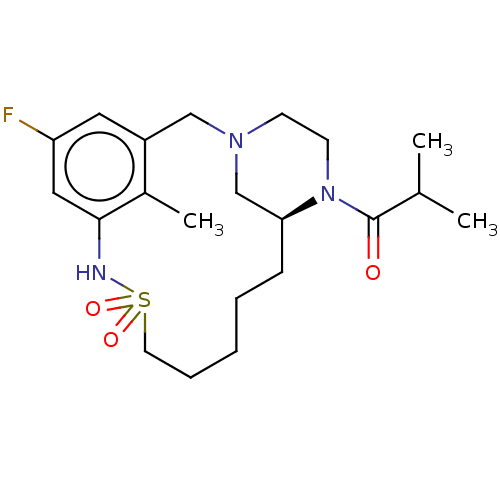
Affinity DataKi: 1.80E+3nMAssay Description:Antagonistic activity against M2 muscarinic receptor in guinea pig left atrium derived by plotting log(DR - 1) vs log[antagonist]More data for this Ligand-Target Pair
In DepthDetails
Affinity DataKi: 3.60E+3nMAssay Description:Antagonistic activity against M2 muscarinic receptor in guinea pig left atrium derived by plotting log(DR - 1) vs log[antagonist]More data for this Ligand-Target Pair
In DepthDetails
Affinity DataKi: 4.00E+3nMAssay Description:Antagonistic activity against M2 muscarinic receptor in guinea pig left atrium derived by plotting log(DR - 1) vs log[antagonist]More data for this Ligand-Target Pair
In DepthDetails
Affinity DataKi: 1.10E+4nMAssay Description:Antagonistic activity against M2 muscarinic receptor in guinea pig left atrium derived by plotting log(DR - 1) vs log[antagonist]More data for this Ligand-Target Pair
In DepthDetails
Affinity DataKi: 1.70E+4nMAssay Description:Antagonistic activity against M2 muscarinic receptor in guinea pig left atrium derived by plotting log(DR - 1) vs log[antagonist]More data for this Ligand-Target Pair
In DepthDetails
Affinity DataKi: 1.90E+4nMAssay Description:Antagonistic activity against M2 muscarinic receptor in guinea pig left atrium derived by plotting log(DR - 1) vs log[antagonist]More data for this Ligand-Target Pair
In DepthDetails
Affinity DataKi: 3.70E+4nMAssay Description:Antagonistic activity against M2 muscarinic receptor in guinea pig left atrium derived by plotting log(DR - 1) vs log[antagonist]More data for this Ligand-Target Pair
In DepthDetails