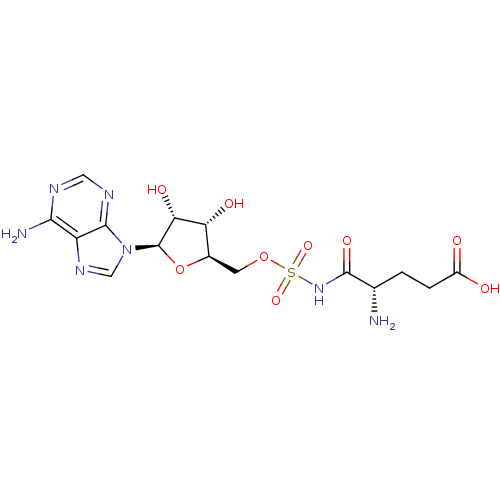
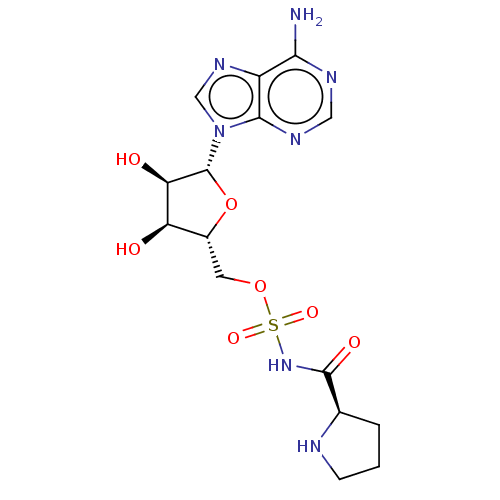
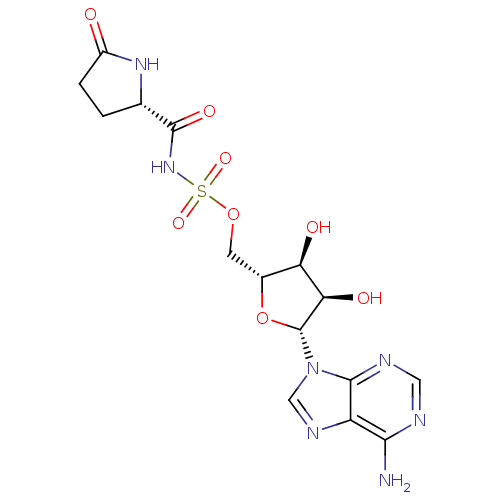
Affinity DataKi: 2.80nM ΔG°: -50.8kJ/molepH: 7.2 T: 2°CAssay Description:The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p...More data for this Ligand-Target Pair
Affinity DataKi: 5.10nMAssay Description:Binding affinity towards 5-hydroxytryptamine 3 receptorMore data for this Ligand-Target Pair
Affinity DataKi: 15nMAssay Description:Binding affinity towards 5-hydroxytryptamine 3 receptorMore data for this Ligand-Target Pair
Affinity DataKi: 70nM ΔG°: -41.5kJ/molepH: 8.0 T: 2°CAssay Description:The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p...More data for this Ligand-Target Pair
Ligand Info
In DepthDetails
Affinity DataKi: 1.50E+4nM ΔG°: -28.6kJ/molepH: 7.2 T: 2°CAssay Description:The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p...More data for this Ligand-Target Pair
Affinity DataKi: 1.80E+4nM ΔG°: -28.2kJ/molepH: 7.2 T: 2°CAssay Description:The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p...More data for this Ligand-Target Pair
Affinity DataKi: 2.90E+6nM ΔG°: -15.1kJ/molepH: 7.2 T: 2°CAssay Description:The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p...More data for this Ligand-Target Pair