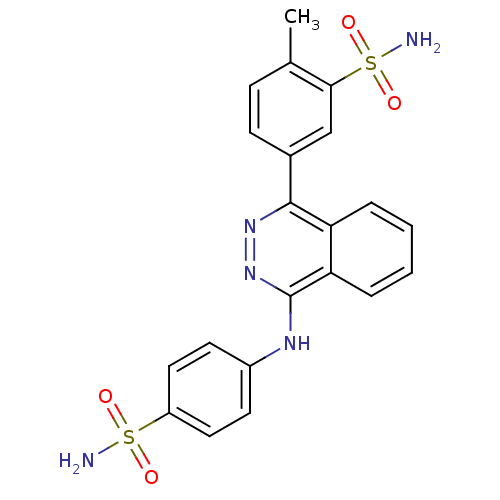
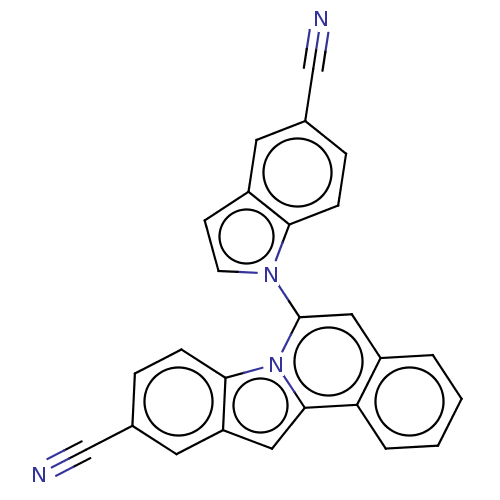
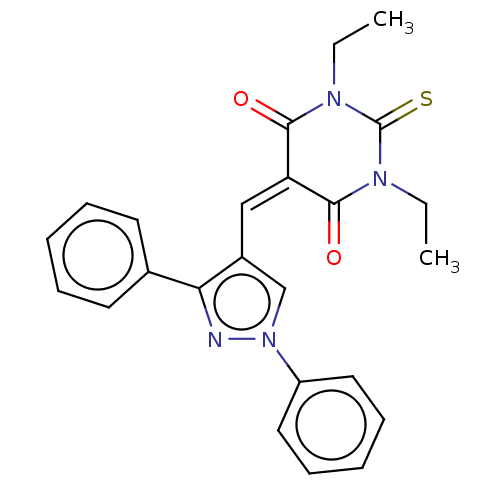
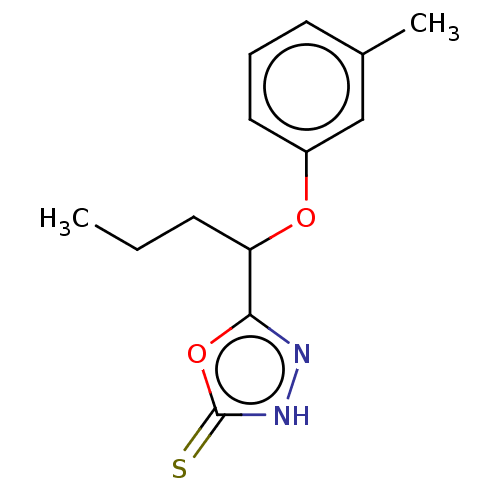
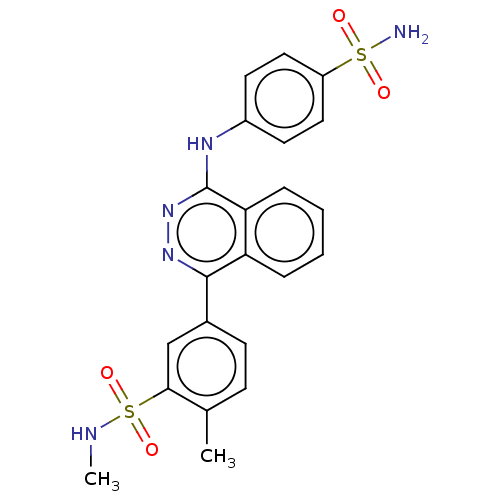
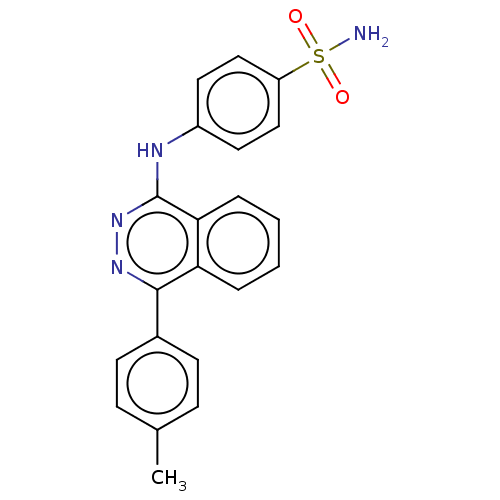
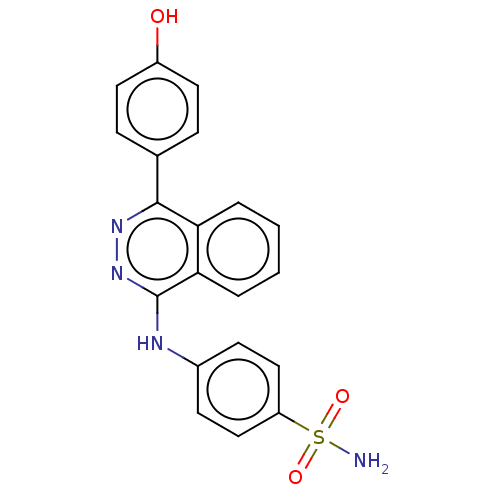
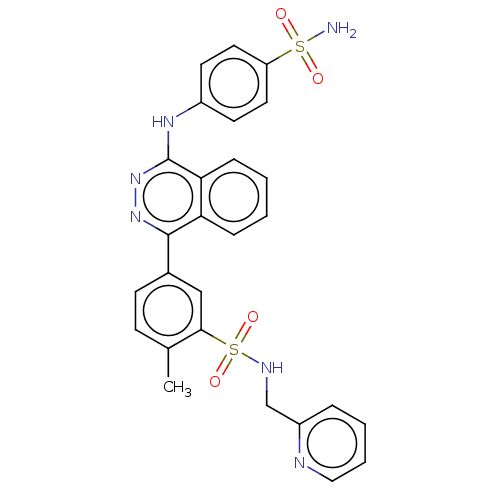
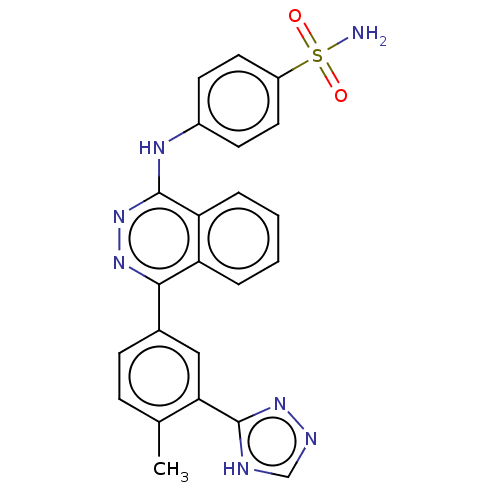
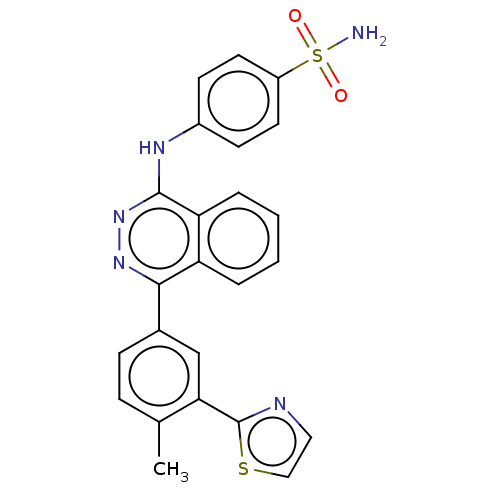
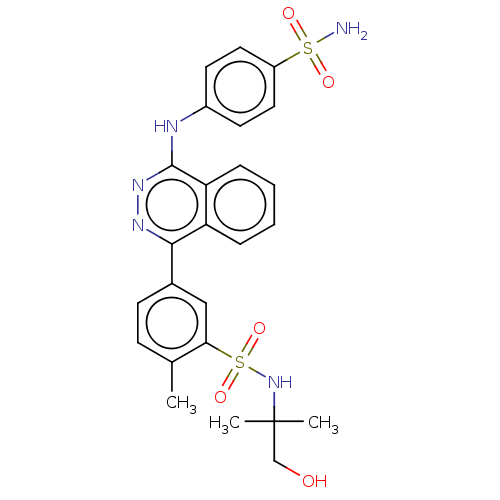
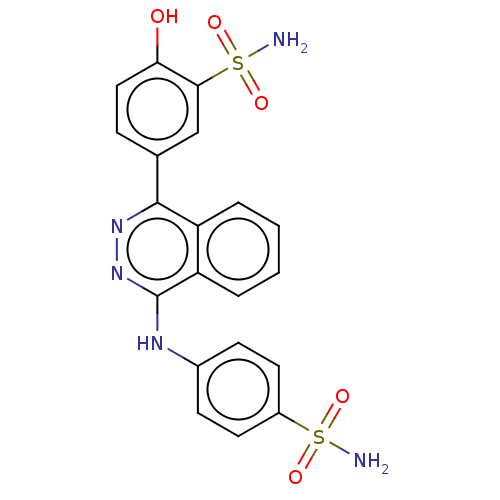
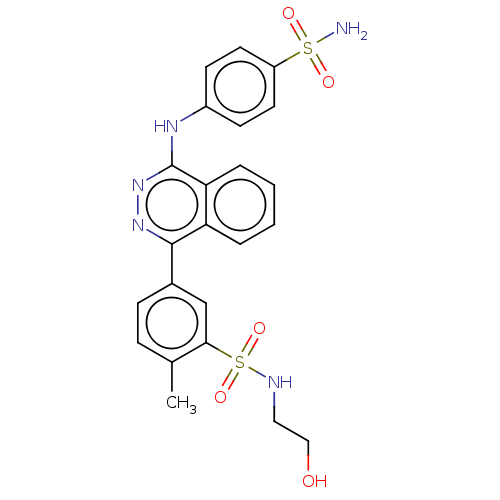
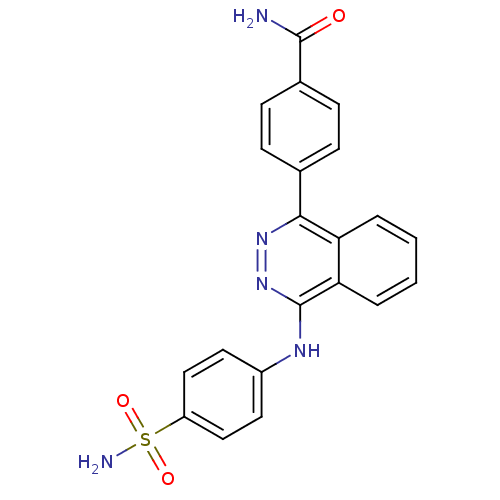
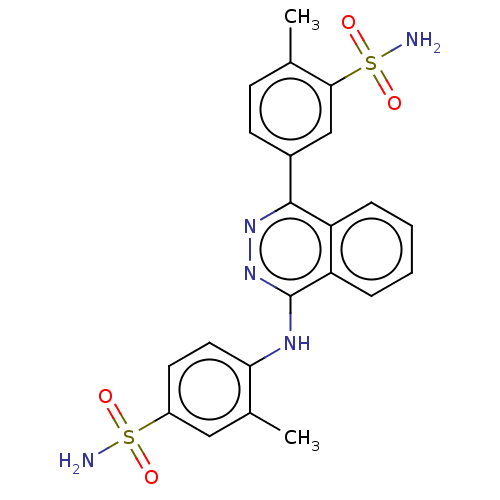
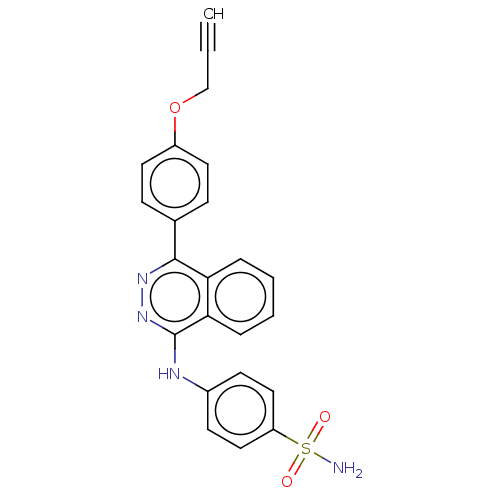
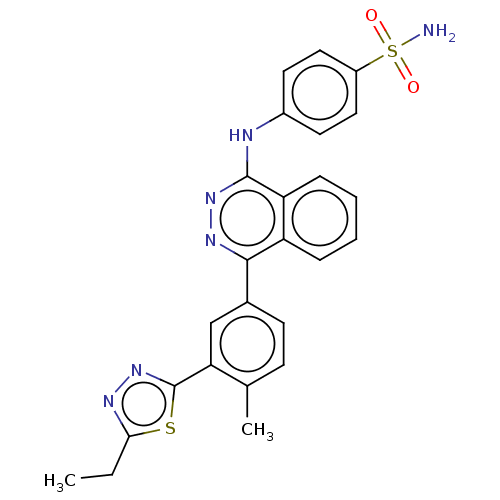
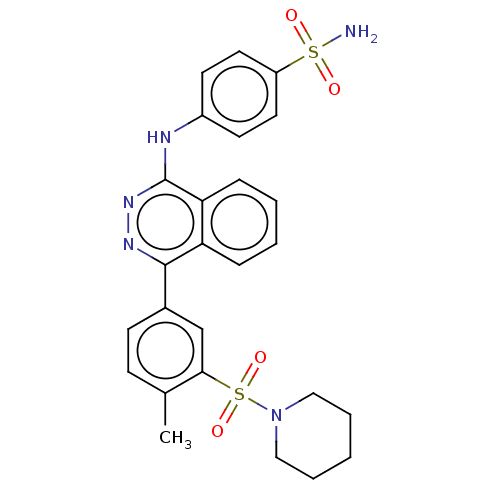
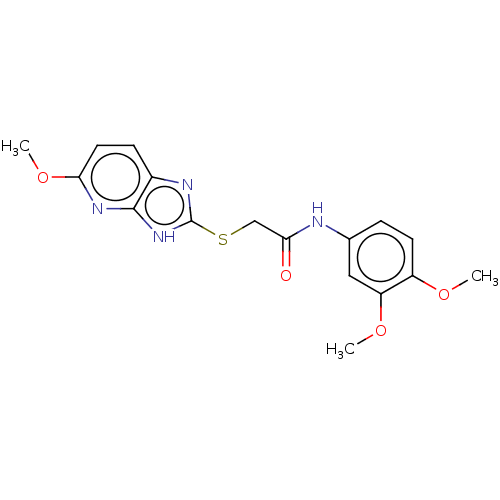
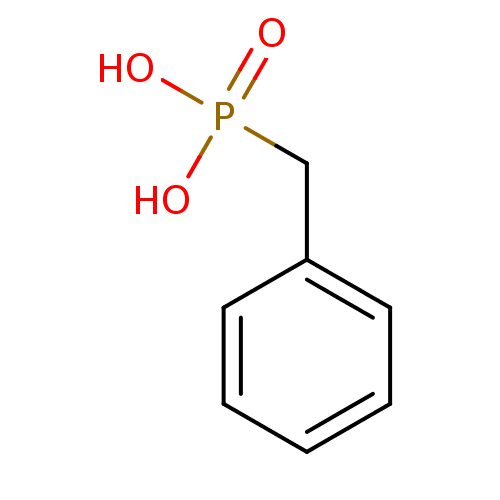
Affinity DataKi: 6.81E+4nMAssay Description:Competitive inhibition of human NPP3 using ATP as substrateMore data for this Ligand-Target Pair
TargetEctonucleotide pyrophosphatase/phosphodiesterase family member 3(Bos taurus)
University of Campinas
Curated by ChEMBL
University of Campinas
Curated by ChEMBL
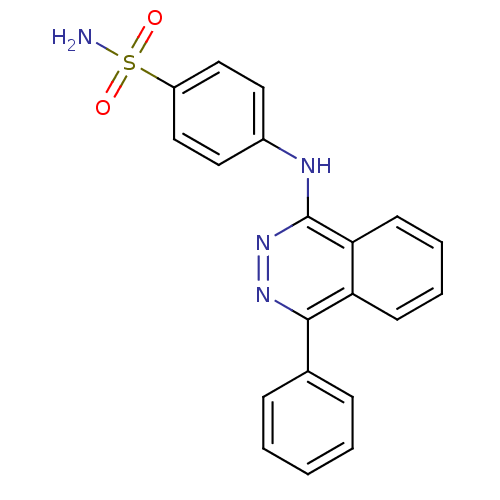
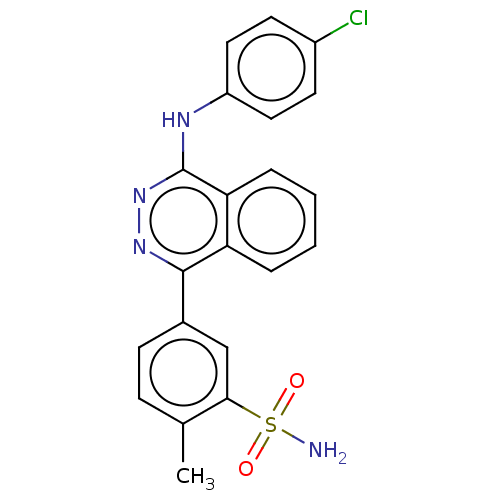
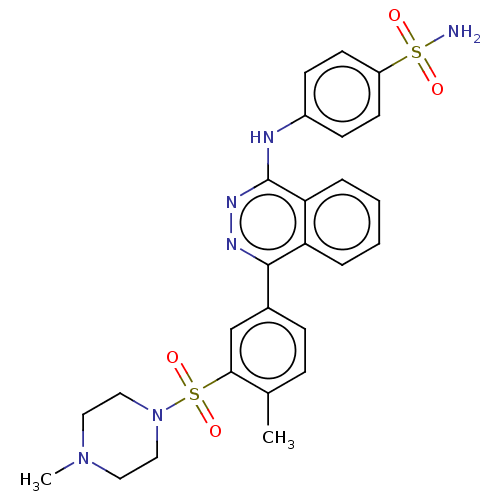
Affinity DataKi: 1.40E+6nMAssay Description:Inhibition of bovine intestinal 5'-nucleotide phosphodiesterase using 4-nitrophenyl phenylphosphonate substrate by spectrophotometryMore data for this Ligand-Target Pair