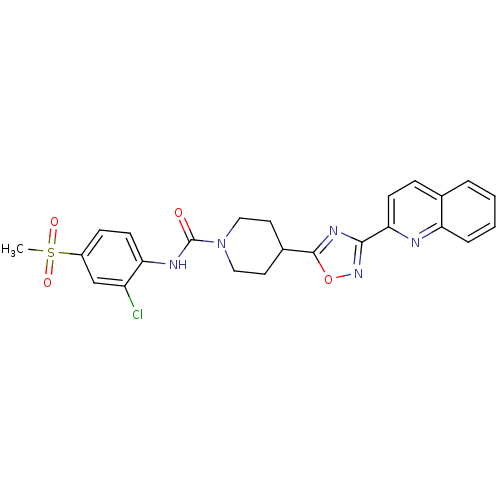
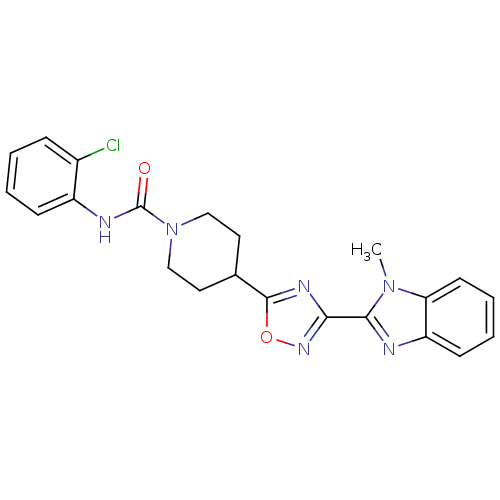
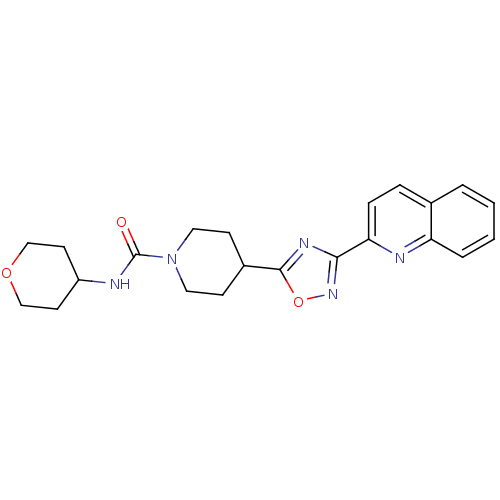
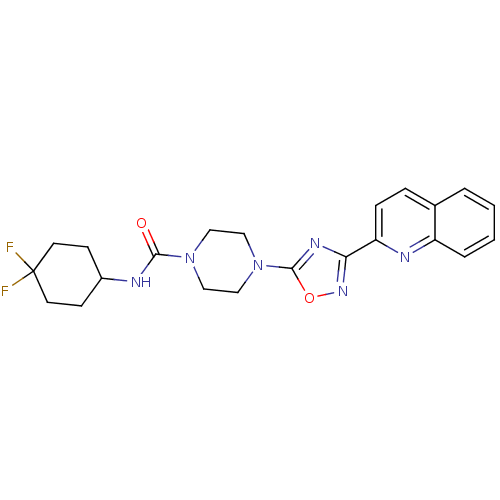
Found 115 hits of Enzyme Inhibition Constant Data
Found 115 hits of Enzyme Inhibition Constant Data
Affinity DataIC50: 3.70E+3nMAssay Description:Inhibition of of Bodipy-labelled cyclopamine binding to Smo expressed in COS-1 cells in presence of 20% NHS after 4 to 6 hrs by FACS flow cytometric ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.10E+3nMAssay Description:Inhibition of of Bodipy-labelled cyclopamine binding to Smo expressed in COS-1 cells in presence of 20% NHS after 4 to 6 hrs by FACS flow cytometric ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.50E+3nMAssay Description:Inhibition of of Bodipy-labelled cyclopamine binding to Smo expressed in COS-1 cells in presence of 20% NHS after 4 to 6 hrs by FACS flow cytometric ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.70E+3nMAssay Description:Displacement of Bodipy-labelled cyclopamine from Smo expressed in COS-1 cells in presence of 2% FBS after 4 to 6 hrs by FACS flow cytometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 4.80E+3nMAssay Description:Inhibition of of Bodipy-labelled cyclopamine binding to Smo expressed in COS-1 cells in presence of 20% NHS after 4 to 6 hrs by FACS flow cytometric ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+3nMAssay Description:Displacement of Bodipy-labelled cyclopamine from Smo expressed in COS-1 cells in presence of 2% FBS after 4 to 6 hrs by FACS flow cytometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 8.60E+3nMAssay Description:Inhibition of of Bodipy-labelled cyclopamine binding to Smo expressed in COS-1 cells in presence of 20% NHS after 4 to 6 hrs by FACS flow cytometric ...More data for this Ligand-Target Pair
TargetTranscriptional regulator ERG(Homo sapiens (Human))
Merck Research Laboratories Rome
Curated by ChEMBL
Merck Research Laboratories Rome
Curated by ChEMBL
Affinity DataIC50: 1.20E+4nMAssay Description:Inhibition of human ERGMore data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+4nMAssay Description:Displacement of Bodipy-labelled cyclopamine from Smo expressed in COS-1 cells in presence of 2% FBS after 4 to 6 hrs by FACS flow cytometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2C9More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP2D6More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of CYP3A4More data for this Ligand-Target Pair
TargetTranscriptional regulator ERG(Homo sapiens (Human))
Merck Research Laboratories Rome
Curated by ChEMBL
Merck Research Laboratories Rome
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human ERGMore data for this Ligand-Target Pair
TargetTranscriptional regulator ERG(Homo sapiens (Human))
Merck Research Laboratories Rome
Curated by ChEMBL
Merck Research Laboratories Rome
Curated by ChEMBL
Affinity DataIC50: 3.00E+4nMAssay Description:Inhibition of human ERGMore data for this Ligand-Target Pair
TargetTranscriptional regulator ERG(Homo sapiens (Human))
Merck Research Laboratories Rome
Curated by ChEMBL
Merck Research Laboratories Rome
Curated by ChEMBL
Affinity DataIC50: 3.20E+4nMAssay Description:Inhibition of human ERGMore data for this Ligand-Target Pair