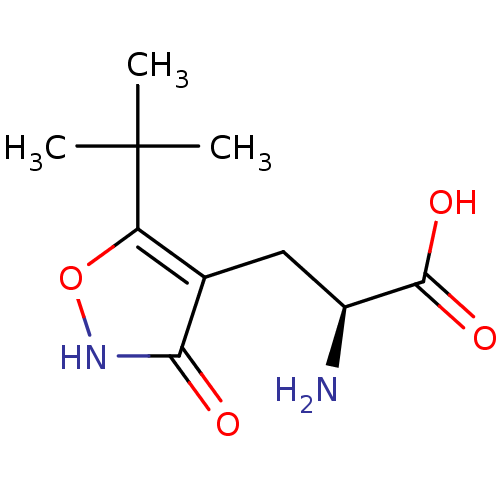
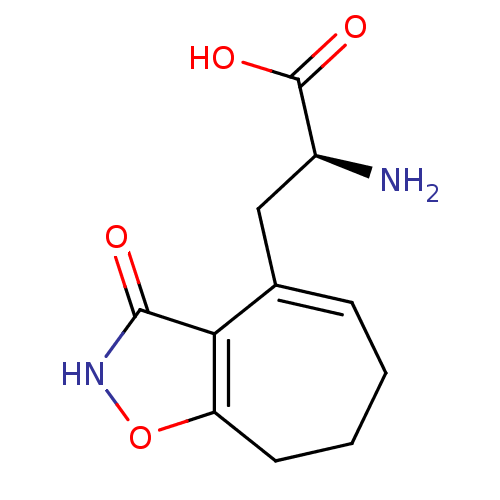
Found 21 hits of Enzyme Inhibition Constant Data
Found 21 hits of Enzyme Inhibition Constant Data
Affinity DataKi: 2.13nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.57nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 5.08nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 173nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 175nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2(R) RNA-edited flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 3(Rattus norvegicus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataKi: 199nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR7A expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 202nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 232nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 1.11E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 3(Rattus norvegicus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataKi: 1.38E+3nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR7A expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 1.55E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 1.66E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2(R) RNA-edited flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.04E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.14E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.20E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 3(Rattus norvegicus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataKi: 2.53E+3nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR7A expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.78E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 6.31E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2(R) RNA-edited flop isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 2(Rattus norvegicus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]KA from rat recombinant GluR6(VCR) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 2(Rattus norvegicus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]KA from rat recombinant GluR6(VCR) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 2(Rattus norvegicus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataKi: >1.00E+6nMAssay Description:Displacement of [3H]KA from rat recombinant GluR6(VCR) RNA-edited isoform expressed in baculovirus infected Sf9 cellsChecked by AuthorMore data for this Ligand-Target Pair