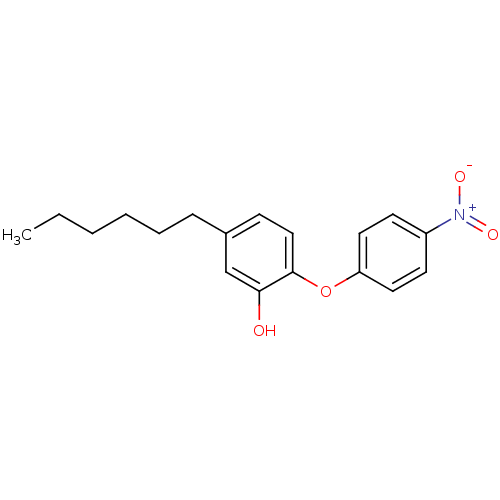
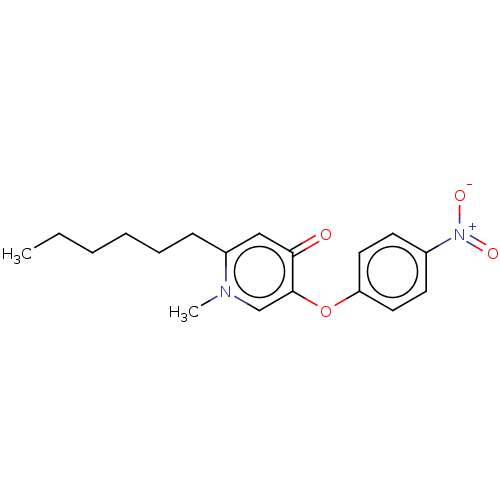
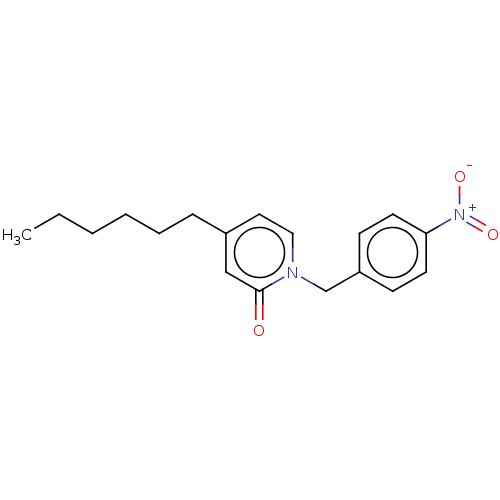
Found 8 hits of Enzyme Inhibition Constant Data
Found 8 hits of Enzyme Inhibition Constant Data
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 100nM ΔG°: -40.0kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 200nM ΔG°: -38.2kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH] [T276S](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 500nM ΔG°: -36.0kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH] [T276S](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 3.00E+3nM ΔG°: -31.5kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH] [T276S](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 4.00E+3nM ΔG°: -30.8kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH] [T276S](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 8.00E+3nM ΔG°: -29.1kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 8.00E+3nM ΔG°: -29.1kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADH](Yersinia pestis (Enterobacteria))
University of Würzburg
University of Würzburg
Affinity DataKi: 1.60E+4nM ΔG°: -27.4kJ/molepH: 8.0 T: 2°CAssay Description:Steady-state kinetics were performed on a Cary 100 Bio (Varian) spectrometer at 25 °C using 30 mM PIPES, 150 mM NaCl, and 1.0 mM EDTA (pH 8.0) as...More data for this Ligand-Target Pair