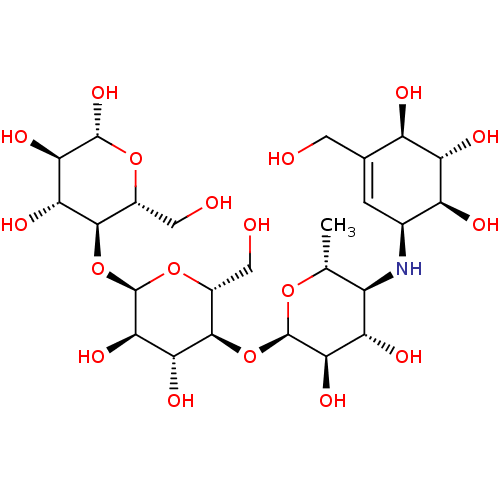
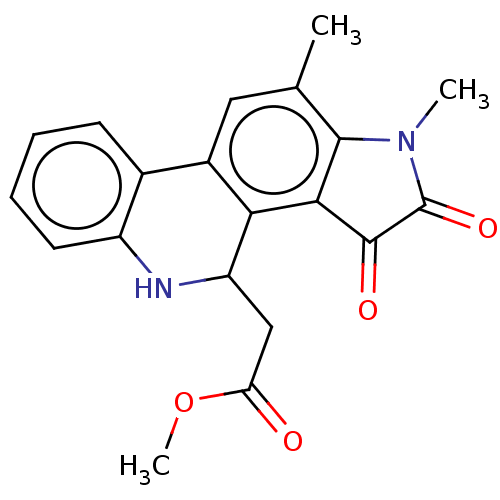
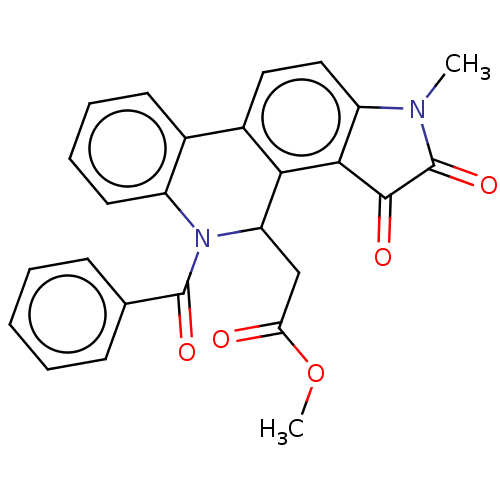
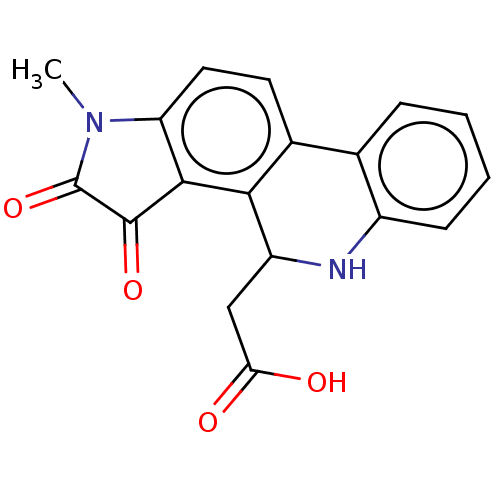
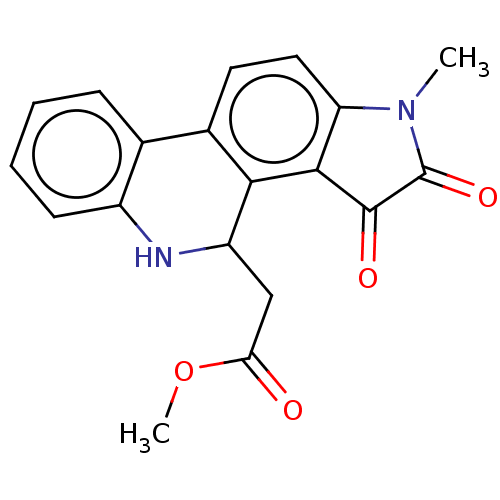
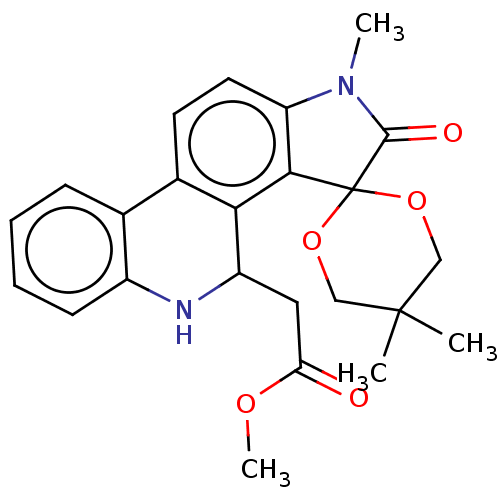
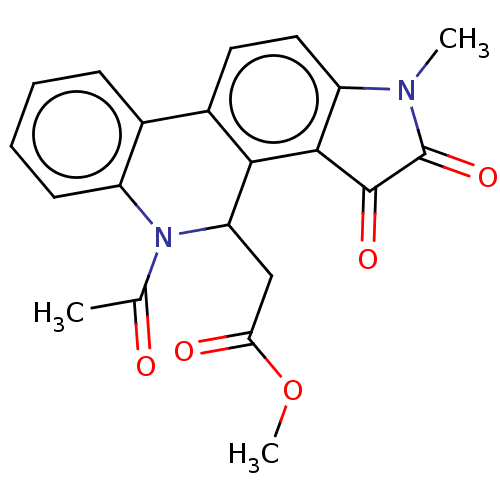
Found 21 hits of Enzyme Inhibition Constant Data
Found 21 hits of Enzyme Inhibition Constant Data
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
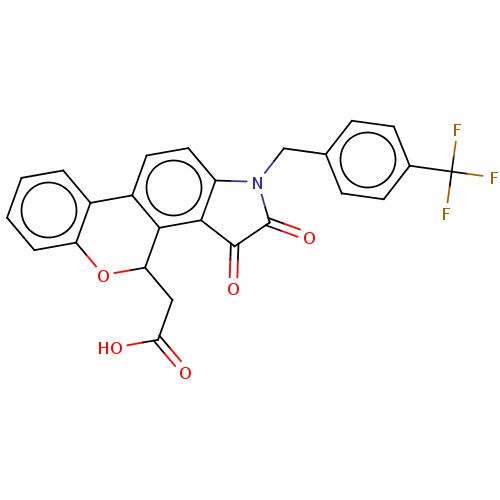
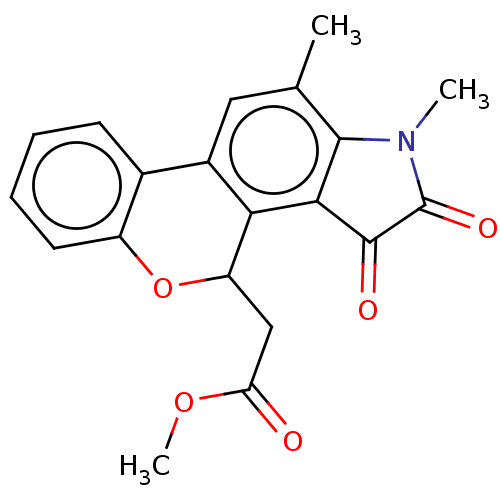
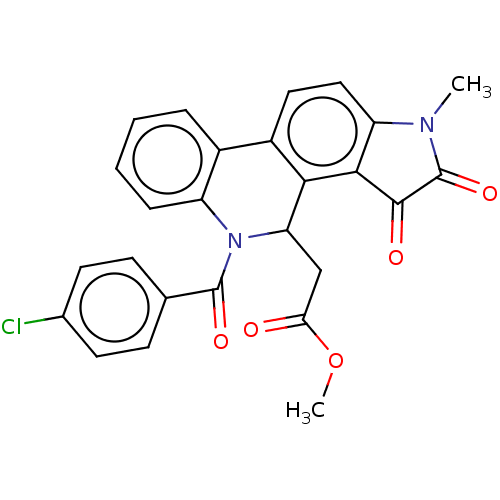
Affinity DataIC50: 4.80E+3nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
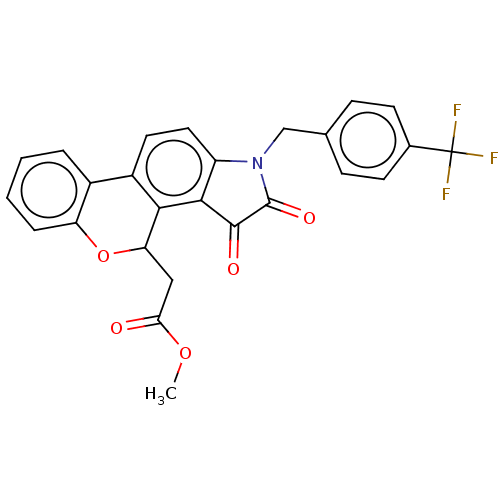
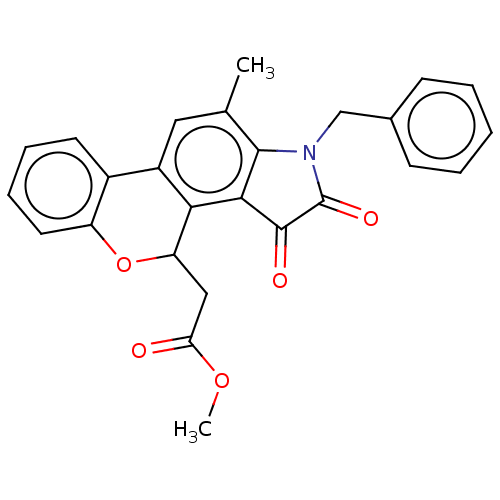
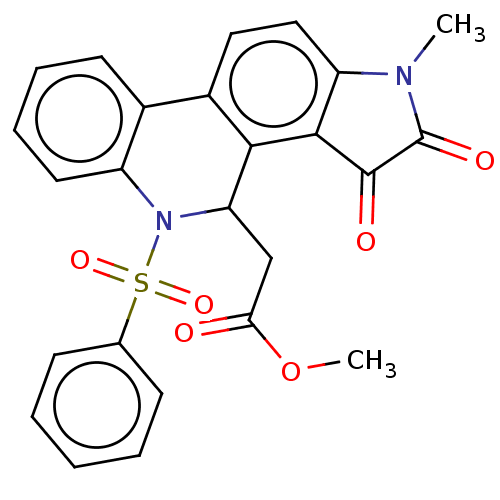
Affinity DataIC50: 1.21E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
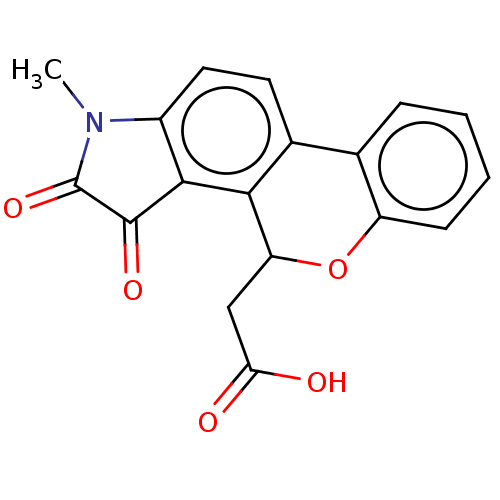
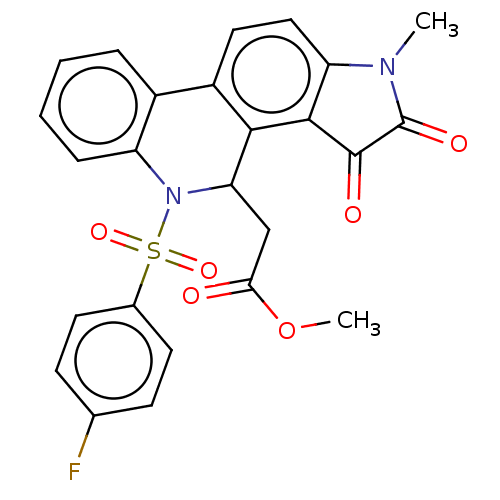
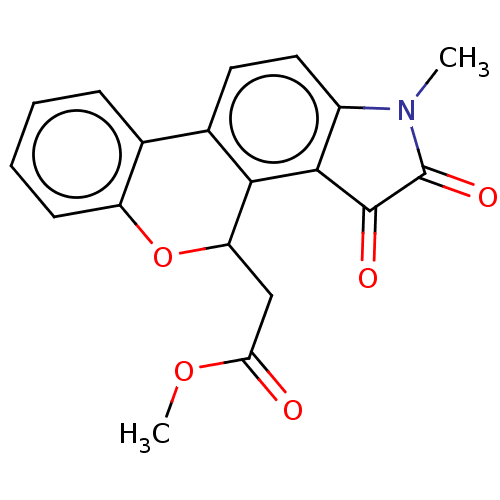
Affinity DataIC50: 1.57E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
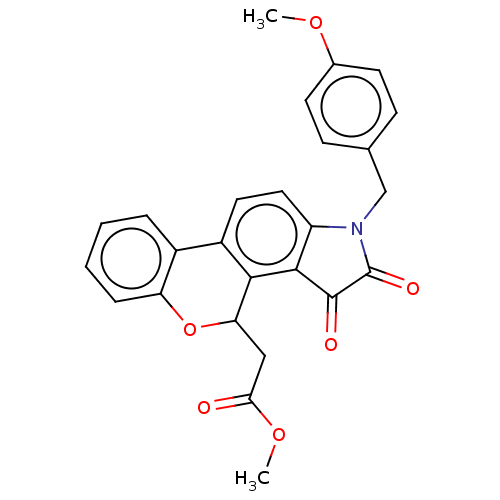
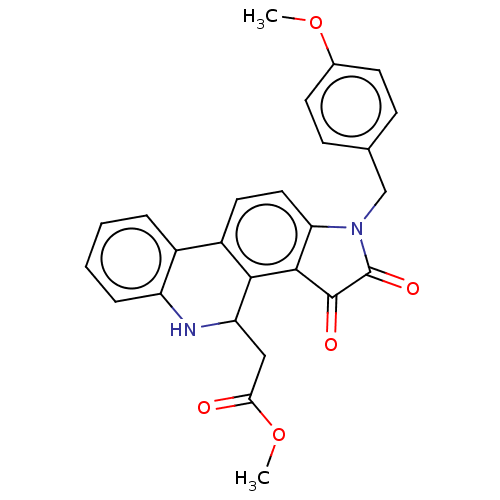
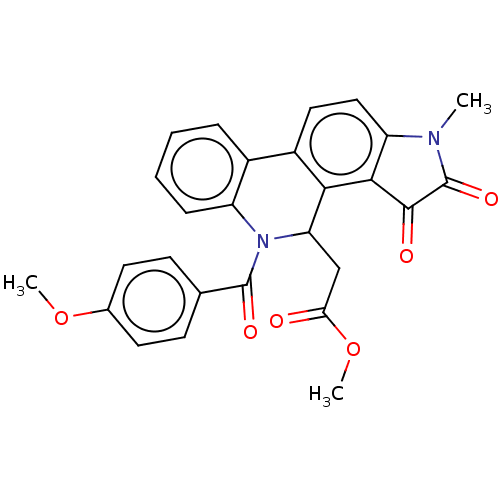
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 2.90E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 3.23E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 3.48E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 3.93E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 4.29E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 4.53E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 7.59E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 7.71E+4nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 1.05E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 1.12E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 1.16E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 1.17E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 1.50E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 1.87E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 2.87E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 3.00E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Key Laboratory of Industrial Fermentation Microbiology (Tianjin University of Science and Technology)
Curated by ChEMBL
Affinity DataIC50: 3.00E+5nMAssay Description:Inhibition of bakers yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate after 30 mins by spectrophotometryMore data for this Ligand-Target Pair