Reaction Details  Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
cAMP-specific 3',5'-cyclic phosphodiesterase 4A
Ligand
BDBM14775
Substrate
n/a
Meas. Tech.
ChEMBL_155727 (CHEMBL760761)
IC50
1.6±n/a nM
Citation
 Kleinman, EF; Campbell, E; Giordano, LA; Cohan, VL; Jenkinson, TH; Cheng, JB; Shirley, JT; Pettipher, ER; Salter, ED; Hibbs, TA; DiCapua, FM; Bordner, J Striking effect of hydroxamic acid substitution on the phosphodiesterase type 4 (PDE4) and TNF alpha inhibitory activity of two series of rolipram analogues: implications for a new active site model of PDE4. J Med Chem 41:266-70 (1998) [PubMed] Article
Kleinman, EF; Campbell, E; Giordano, LA; Cohan, VL; Jenkinson, TH; Cheng, JB; Shirley, JT; Pettipher, ER; Salter, ED; Hibbs, TA; DiCapua, FM; Bordner, J Striking effect of hydroxamic acid substitution on the phosphodiesterase type 4 (PDE4) and TNF alpha inhibitory activity of two series of rolipram analogues: implications for a new active site model of PDE4. J Med Chem 41:266-70 (1998) [PubMed] ArticleMore Info.:
Target
Name:
cAMP-specific 3',5'-cyclic phosphodiesterase 4A
Synonyms:
3',5'-cyclic phosphodiesterase | DPDE2 | PDE46 | PDE4A | PDE4A_HUMAN | Phosphodiesterase 4 (PDE4) | Phosphodiesterase 4A | Phosphodiesterase 4A (PDE4)
Type:
Enzyme
Mol. Mass.:
98113.27
Organism:
Homo sapiens (Human)
Description:
P27815
Residue:
886
Sequence:
MEPPTVPSERSLSLSLPGPREGQATLKPPPQHLWRQPRTPIRIQQRGYSDSAERAERERQPHRPIERADAMDTSDRPGLRTTRMSWPSSFHGTGTGSGGAGGGSSRRFEAENGPTPSPGRSPLDSQASPGLVLHAGAATSQRRESFLYRSDSDYDMSPKTMSRNSSVTSEAHAEDLIVTPFAQVLASLRSVRSNFSLLTNVPVPSNKRSPLGGPTPVCKATLSEETCQQLARETLEELDWCLEQLETMQTYRSVSEMASHKFKRMLNRELTHLSEMSRSGNQVSEYISTTFLDKQNEVEIPSPTMKEREKQQAPRPRPSQPPPPPVPHLQPMSQITGLKKLMHSNSLNNSNIPRFGVKTDQEELLAQELENLNKWGLNIFCVSDYAGGRSLTCIMYMIFQERDLLKKFRIPVDTMVTYMLTLEDHYHADVAYHNSLHAADVLQSTHVLLATPALDAVFTDLEILAALFAAAIHDVDHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQEDNCDIFQNLSKRQRQSLRKMVIDMVLATDMSKHMTLLADLKTMVETKKVTSSGVLLLDNYSDRIQVLRNMVHCADLSNPTKPLELYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTASVEKSQVGFIDYIVHPLWETWADLVHPDAQEILDTLEDNRDWYYSAIRQSPSPPPEEESRGPGHPPLPDKFQFELTLEEEEEEEISMAQIPCTAQEALTAQGLSGVEEALDATIAWEASPAQESLEVMAQEASLEAELEAVYLTQQAQSTGSAPVAPDEFSSREEFVVAVSHSSPSALALQSPLLPAWRTLSVSEHAPGLPGLPSTAAEVEAQREHQAAKRACSACAGTFGEDTSALPAPGGGGSGGDPT
Inhibitor
Name:
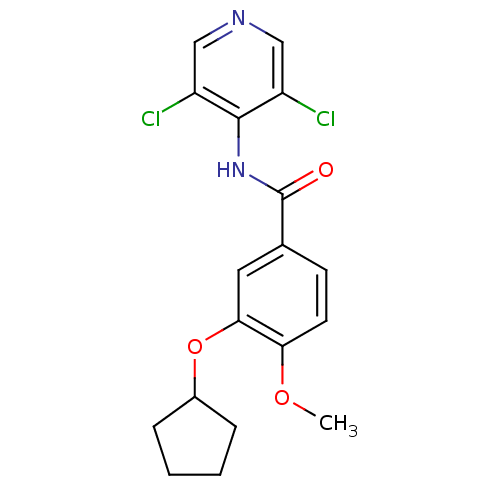
BDBM14775
Synonyms:
3-(cyclopentyloxy)-N-(3,5-dichloropyridin-4-yl)-4-methoxybenzamide | 3-cyclopentyloxy-N-(3,5-dichloropyridin-4-yl)-4-methoxy-benzamide | CHEMBL42126 | Cpodpmb | Piclamilast | RPR-73401
Type:
Small organic molecule
Emp. Form.:
C18H18Cl2N2O3
Mol. Mass.:
381.253
SMILES:
COc1ccc(cc1OC1CCCC1)C(=O)Nc1c(Cl)cncc1Cl