Reaction Details  Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Matrix metalloproteinase-9
Ligand
BDBM50104978
Substrate
n/a
Meas. Tech.
ChEMBL_105383 (CHEMBL872557)
Ki
91±n/a nM
Citation
 Xue, CB; He, X; Corbett, RL; Roderick, J; Wasserman, ZR; Liu, RQ; Jaffee, BD; Covington, MB; Qian, M; Trzaskos, JM; Newton, RC; Magolda, RL; Wexler, RR; Decicco, CP Discovery of macrocyclic hydroxamic acids containing biphenylmethyl derivatives at P1', a series of selective TNF-alpha converting enzyme inhibitors with potent cellular activity in the inhibition of TNF-alpha release. J Med Chem 44:3351-4 (2001) [PubMed]
Xue, CB; He, X; Corbett, RL; Roderick, J; Wasserman, ZR; Liu, RQ; Jaffee, BD; Covington, MB; Qian, M; Trzaskos, JM; Newton, RC; Magolda, RL; Wexler, RR; Decicco, CP Discovery of macrocyclic hydroxamic acids containing biphenylmethyl derivatives at P1', a series of selective TNF-alpha converting enzyme inhibitors with potent cellular activity in the inhibition of TNF-alpha release. J Med Chem 44:3351-4 (2001) [PubMed]More Info.:
Target
Name:
Matrix metalloproteinase-9
Synonyms:
67 kDa matrix metalloproteinase-9 | 82 kDa matrix metalloproteinase-9 | 92 kDa gelatinase | 92 kDa type IV collagenase | CLG4B | GELB | Gelatinase B | MMP-9 | MMP9 | MMP9_HUMAN | Matrix metalloproteinase 9 (MMP-9) | Matrix metalloproteinase-9 (MMP-9) | Matrix metalloproteinase-9 (MMP9)
Type:
Enzyme
Mol. Mass.:
78452.28
Organism:
Homo sapiens (Human)
Description:
P14780
Residue:
707
Sequence:
MSLWQPLVLVLLVLGCCFAAPRQRQSTLVLFPGDLRTNLTDRQLAEEYLYRYGYTRVAEMRGESKSLGPALLLLQKQLSLPETGELDSATLKAMRTPRCGVPDLGRFQTFEGDLKWHHHNITYWIQNYSEDLPRAVIDDAFARAFALWSAVTPLTFTRVYSRDADIVIQFGVAEHGDGYPFDGKDGLLAHAFPPGPGIQGDAHFDDDELWSLGKGVVVPTRFGNADGAACHFPFIFEGRSYSACTTDGRSDGLPWCSTTANYDTDDRFGFCPSERLYTQDGNADGKPCQFPFIFQGQSYSACTTDGRSDGYRWCATTANYDRDKLFGFCPTRADSTVMGGNSAGELCVFPFTFLGKEYSTCTSEGRGDGRLWCATTSNFDSDKKWGFCPDQGYSLFLVAAHEFGHALGLDHSSVPEALMYPMYRFTEGPPLHKDDVNGIRHLYGPRPEPEPRPPTTTTPQPTAPPTVCPTGPPTVHPSERPTAGPTGPPSAGPTGPPTAGPSTATTVPLSPVDDACNVNIFDAIAEIGNQLYLFKDGKYWRFSEGRGSRPQGPFLIADKWPALPRKLDSVFEERLSKKLFFFSGRQVWVYTGASVLGPRRLDKLGLGADVAQVTGALRSGRGKMLLFSGRRLWRFDVKAQMVDPRSASEVDRMFPGVPLDTHDVFQYREKAYFCQDRFYWRVSSRSELNQVDQVGYVTYDILQCPED
Inhibitor
Name:
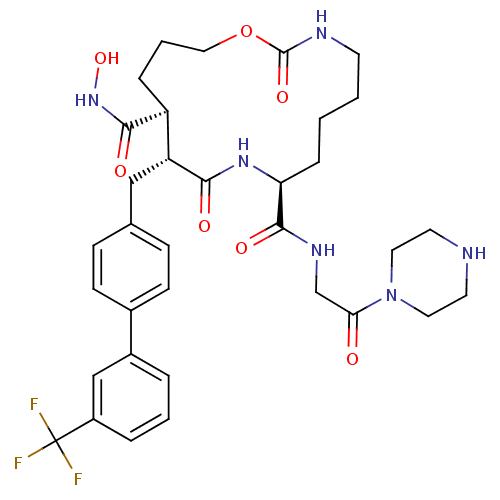
BDBM50104978
Synonyms:
2,10-Dioxo-11-(3'-trifluoromethyl-biphenyl-4-ylmethyl)-1-oxa-3,9-diaza-cyclopentadecane-8,12-dicarboxylic acid 12-hydroxyamide 8-[(2-oxo-2-piperazin-1-yl-ethyl)-amide] | CHEMBL325421
Type:
Small organic molecule
Emp. Form.:
C34H43F3N6O7
Mol. Mass.:
704.7364
SMILES:
ONC(=O)[C@H]1CCCOC(=O)NCCCC[C@H](NC(=O)[C@@H]1Cc1ccc(cc1)-c1cccc(c1)C(F)(F)F)C(=O)NCC(=O)N1CCNCC1