Reaction Details  Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Glutamate receptor ionotropic, kainate 2
Ligand
BDBM50116050
Substrate
n/a
Meas. Tech.
ChEBML_72854
Ki
5800±n/a nM
Citation
 Lubisch, W; Behl, B; Henn, C; Hofmann, HP; Reeb, J; Regner, F; Vierling, M Pyrrolylquinoxalinediones carrying a piperazine residue represent highly potent and selective ligands to the homomeric kainate receptor GluR5. Bioorg Med Chem Lett 12:2113-6 (2002) [PubMed]
Lubisch, W; Behl, B; Henn, C; Hofmann, HP; Reeb, J; Regner, F; Vierling, M Pyrrolylquinoxalinediones carrying a piperazine residue represent highly potent and selective ligands to the homomeric kainate receptor GluR5. Bioorg Med Chem Lett 12:2113-6 (2002) [PubMed]More Info.:
Target
Name:
Glutamate receptor ionotropic, kainate 2
Synonyms:
EAA4 | Excitatory amino acid receptor 4 | GLUR6 | GRIK2 | GRIK2_HUMAN | GluK2 | GluR-6 | Glutamate kainate | Glutamate receptor 6 | Glutamate receptor ionotropic kainate 2 | Glutamate receptor, ionotropic kainate 2 | Glutamate-Kainate | Glutamate-Kainate, GluR6
Type:
Enzyme Catalytic Domain
Mol. Mass.:
102592.78
Organism:
Homo sapiens (Human)
Description:
Q13002
Residue:
908
Sequence:
MKIIFPILSNPVFRRTVKLLLCLLWIGYSQGTTHVLRFGGIFEYVESGPMGAEELAFRFAVNTINRNRTLLPNTTLTYDTQKINLYDSFEASKKACDQLSLGVAAIFGPSHSSSANAVQSICNALGVPHIQTRWKHQVSDNKDSFYVSLYPDFSSLSRAILDLVQFFKWKTVTVVYDDSTGLIRLQELIKAPSRYNLRLKIRQLPADTKDAKPLLKEMKRGKEFHVIFDCSHEMAAGILKQALAMGMMTEYYHYIFTTLDLFALDVEPYRYSGVNMTGFRILNTENTQVSSIIEKWSMERLQAPPKPDSGLLDGFMTTDAALMYDAVHVVSVAVQQFPQMTVSSLQCNRHKPWRFGTRFMSLIKEAHWEGLTGRITFNKTNGLRTDFDLDVISLKEEGLEKIGTWDPASGLNMTESQKGKPANITDSLSNRSLIVTTILEEPYVLFKKSDKPLYGNDRFEGYCIDLLRELSTILGFTYEIRLVEDGKYGAQDDANGQWNGMVRELIDHKADLAVAPLAITYVREKVIDFSKPFMTLGISILYRKPNGTNPGVFSFLNPLSPDIWMYILLAYLGVSCVLFVIARFSPYEWYNPHPCNPDSDVVENNFTLLNSFWFGVGALMQQGSELMPKALSTRIVGGIWWFFTLIIISSYTANLAAFLTVERMESPIDSADDLAKQTKIEYGAVEDGATMTFFKKSKISTYDKMWAFMSSRRQSVLVKSNEEGIQRVLTSDYAFLMESTTIEFVTQRNCNLTQIGGLIDSKGYGVGTPMGSPYRDKITIAILQLQEEGKLHMMKEKWWRGNGCPEEESKEASALGVQNIGGIFIVLAAGLVLSVFVAVGEFLYKSKKNAQLEKRSFCSAMVEELRMSLKCQRRLKHKPQAPVIVKTEEVINMHTFNDRRLPGKETMA
Inhibitor
Name:
BDBM50116050
Synonyms:
1-(4-Carboxymethyl-7-nitro-2,3-dioxo-1,2,3,4-tetrahydro-quinoxalin-6-yl)-1H-pyrrole-3-carboxylic acid | CHEMBL293613
Type:
Small organic molecule
Emp. Form.:
C15H10N4O8
Mol. Mass.:
374.2619
SMILES:
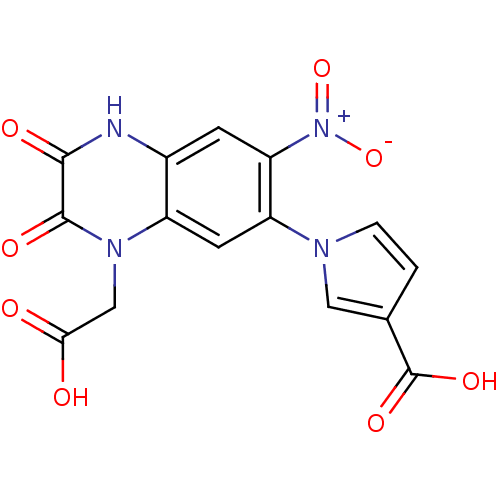
OC(=O)Cn1c2cc(c(cc2[nH]c(=O)c1=O)[N+]([O-])=O)-n1ccc(c1)C(O)=O