Reaction Details  Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Trifunctional purine biosynthetic protein adenosine-3
Ligand
BDBM50016660
Substrate
n/a
Meas. Tech.
ChEBML_70037
IC50
>0.020±n/a nM
Comments
>0.02 20220121
Citation
 Nair, MG; Murthy, BR; Patil, SD; Kisliuk, RL; Thorndike, J; Gaumont, Y; Ferone, R; Duch, DS; Edelstein, MP Folate analogues. 31. Synthesis of the reduced derivatives of 11-deazahomofolic acid, 10-methyl-11-deazahomofolic acid, and their evaluation as inhibitors of glycinamide ribonucleotide formyltransferase. J Med Chem 32:1277-83 (1989) [PubMed]
Nair, MG; Murthy, BR; Patil, SD; Kisliuk, RL; Thorndike, J; Gaumont, Y; Ferone, R; Duch, DS; Edelstein, MP Folate analogues. 31. Synthesis of the reduced derivatives of 11-deazahomofolic acid, 10-methyl-11-deazahomofolic acid, and their evaluation as inhibitors of glycinamide ribonucleotide formyltransferase. J Med Chem 32:1277-83 (1989) [PubMed]More Info.:
Target
Name:
Trifunctional purine biosynthetic protein adenosine-3
Synonyms:
GAR transformylase | Gart | PUR2_MOUSE
Type:
PROTEIN
Mol. Mass.:
107504.27
Organism:
Mus musculus
Description:
ChEMBL_497843
Residue:
1010
Sequence:
MAARVLVIGSGGREHTLAWKLAQSPQVKQVLVAPGNAGTACAGKISNAAVSVNDHSALAQFCKDEKIELVVVGPEAPLAAGIVGDLTSAGVRCFGPTAQAAQLESSKKFAKEFMDRHEIPTAQWRAFTNPEDACSFITSANFPALVVKASGLAAGKGVIVAKSQAEACRAVQEIMQEKSFGAAGETVVVEEFLEGEEVSCLCFTDGKTVAEMPPAQDHKRLLDGDEGPNTGGMGAYCPAPQVSKDLLVKIKNTILQRAVDGMQQEGAPYTGILYAGIMLTKDGPKVLEFNCRFGDPECQVILPLLKSDLYEVMQSTLDGLLSASLPVWLENHSAVTVVMASKGYPGAYTKGVEITGFPEAQALGLQVFHAGTALKDGKVVTSGGRVLTVTAVQENLMSALAEARKGLAALKFEGAIYRKDIGFRAVAFLQRPRGLTYKDSGVDIAAGNMLVKKIQPLAKATSRPGCSVDLGGFAGLFDLKAAGFKDPLLASGTDGVGTKLKIAQLCNKHDSIGQDLVAMCVNDILAQGAEPLFFLDYFSCGKLDLSTTEAVIAGIAAACQQAGCALLGGETAEMPNMYPPGEYDLAGFAVGAMERHQKLPQLERITEGDAVIGVASSGLHSNGFSLVRKIVERSSLQYSSPAPGGCGDQTLGDLLLTPTRIYSHSLLPIIRSGRVKAFAHITGGGLLENIPRVLPQKFGVDLDASTWRVPKVFSWLQQEGELSEEEMARTFNCGIGAALVVSKDQAEQVLHDVRRRQEEAWVIGSVVACPEDSPRVRVKNLIETIQTNGSLVANGFLKSNFPVQQKKARVAVLISGTGSNLQALIDSTRDPKSSSHIVLVISNKAAVAGLDRAERAGIPTRVINHKLYKNRVEFDNAVDHVLEEFSVDIVCLAGFMRILSGPFVRKWDGKMLNIHPSLLPSFKGSNAHEQVLEAGVTITGCTVHFVAEDVDAGQIILQEAVPVRRGDTVATLSERVKVAEHKIFPAALQLVASGAVQLREDGKIHWAKEQ
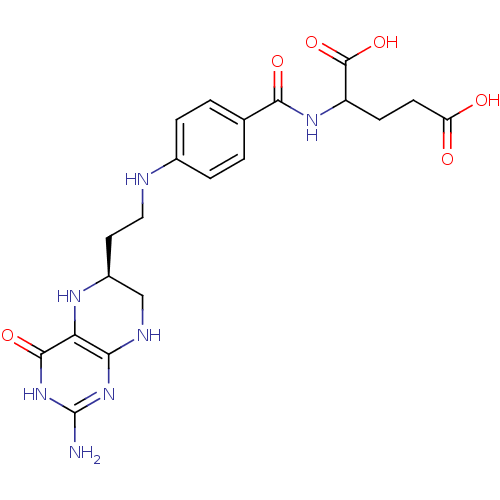
Inhibitor
Name:
BDBM50016660
Synonyms:
2-{4-[2-(2-Amino-4-oxo-3,4,5,6,7,8-hexahydro-pteridin-6-yl)-ethylamino]-benzoylamino}-pentanedioic acid | CHEMBL23388
Type:
Small organic molecule
Emp. Form.:
C20H25N7O6
Mol. Mass.:
459.4558
SMILES:
Nc1nc2NC[C@H](CCNc3ccc(cc3)C(=O)NC(CCC(O)=O)C(O)=O)Nc2c(=O)[nH]1