

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM15695 N-(3-chlorophenyl)-5-oxo-1-phenylpyrrolidine-3-carboxamide::Pyrrolidine Carboxamide Compound 3a
SMILES: Clc1cccc(NC(=O)C2CN(C(=O)C2)c2ccccc2)c1
InChI Key: InChIKey=CMNQHDYAXMXRDF-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enoyl-ACP Reductase (InhA) (Mycobacterium tuberculosis (strain ATCC 25618 / H3...) | BDBM15695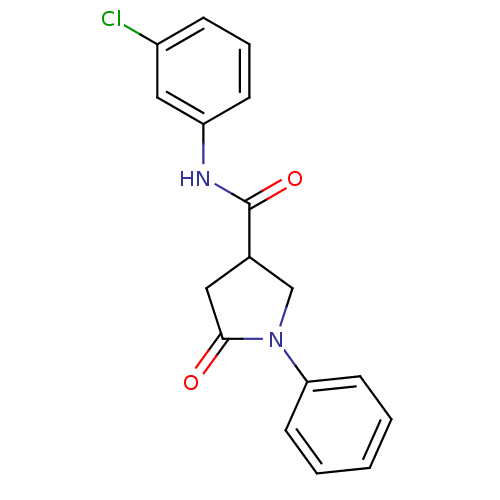 (N-(3-chlorophenyl)-5-oxo-1-phenylpyrrolidine-3-car...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.94E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of California at San Francisco | Assay Description The assay measured the NADH-dependent catalysis of an octenoyl-CoA substrate as a decrease in 340 nm absorbance resulting from conversion of NADH to ... | J Med Chem 49: 6308-23 (2006) Article DOI: 10.1021/jm060715y BindingDB Entry DOI: 10.7270/Q2707ZP0 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||