

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: Cn1cc(c(n1)-c1ccc(OCc2nc3ccccc3n2C)cc1)-c1ccncc1
InChI Key: InChIKey=FHQWSFMPRITQCW-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (Rattus norvegicus (rat)) | BDBM31605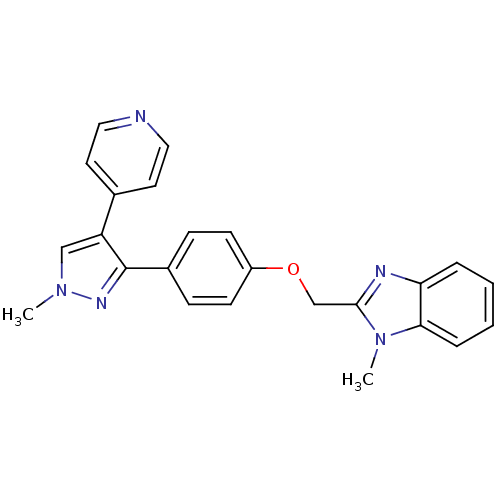 (methyl substituted pyrazole, 26) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 1.29 | n/a | n/a | n/a | n/a | 7.5 | 23 |
Pfizer | Assay Description PDE activity was measured using a plate-based Scintillation Proximity Assay (SPA). For competitive enzyme inhibition assays, the substrate [3H]cAMP c... | J Med Chem 52: 5188-96 (2009) Article DOI: 10.1021/jm900521k BindingDB Entry DOI: 10.7270/Q290223B | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||