

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: CS(=O)(=O)CCNC(=O)c1ccc(cc1F)N1C[C@@H]2C[C@H]1C[C@H]2OCc1c(onc1-c1c(Cl)cccc1Cl)C1CC1
InChI Key: InChIKey=GZKPDTSPKGWKDB-FXJWSJNKSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bile acid receptor (Homo sapiens (Human)) | BDBM465521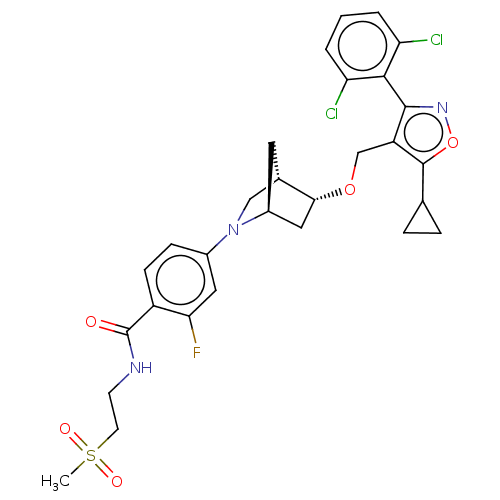 (US10793568, Compound I-170) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | US Patent | n/a | n/a | n/a | n/a | 0.398 | n/a | n/a | n/a | n/a |
ARDELYX, INC. US Patent | Assay Description The affinity of FXR ligands for the ligand binding domain of FXR was determined using a commercially available human FXR ligand binding assay (Lantha... | US Patent US10793568 (2020) BindingDB Entry DOI: 10.7270/Q21C20Z1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||