

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM4690 (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S,3R)-4-[2H-1,3-benzodioxole-5-(2-methylpropyl)sulfonamido]-3-hydroxy-1-[4-(pyridin-2-ylmethoxy)phenyl]butan-2-yl]carbamate::GW0385 analog 5
SMILES: [H][C@@]12CCO[C@]1([H])OC[C@@H]2OC(=O)N[C@@H](Cc1ccc(OCc2ccccn2)cc1)[C@H](O)CN(CC(C)C)S(=O)(=O)c1ccc2OCOc2c1
InChI Key: InChIKey=CFXFLQMJJDCVNR-MSUBEGKCSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 Protease (Human immunodeficiency virus type 1) | BDBM4690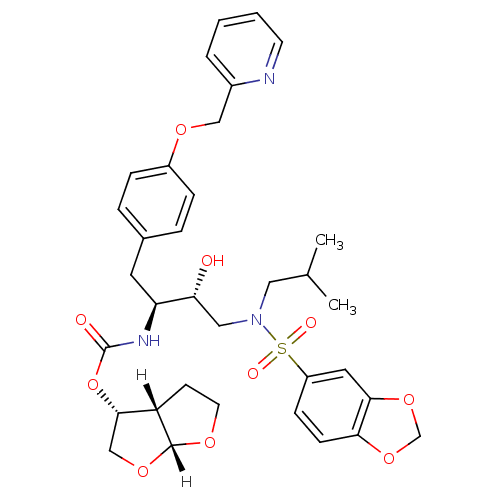 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00000600 | -19.7 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Human immunodeficiency virus type 1 protease (Human immunodeficiency virus type 1) | BDBM4690 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/TrEMBL B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00000600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline Curated by ChEMBL | Assay Description Displacement of [3H]GW0385 from HIV1 protease | Bioorg Med Chem Lett 16: 1788-94 (2006) Article DOI: 10.1016/j.bmcl.2006.01.035 BindingDB Entry DOI: 10.7270/Q2WS8V1F | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 Protease Mutant (M46I,L63P,A71V,V82F,I84V) (Human immunodeficiency virus type 1) | BDBM4690 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.000420 | -17.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 Protease Mutant (L10I,L19Q,K20R,E35D,M36I, ..) (Human immunodeficiency virus type 1) | BDBM4690 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00170 | -16.3 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 Protease Mutant (I50V) (Human immunodeficiency virus type 1) | BDBM4690 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00260 | -16.1 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||