

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: Nc1ncnc2n(cnc12)[C@@H]1O[C@H](COP(O)(=O)O[C@@H]2[C@H](O)[C@@H](COP(O)(=O)O[C@@H]3[C@H](O)[C@@H](COP(O)(=O)OP(O)(=O)OP(O)(O)=O)O[C@H]3n3cnc4c(N)ncnc34)O[C@H]2n2cnc3c(N)ncnc23)[C@@H](O)[C@H]1O
InChI Key: InChIKey=RTAGLZBJCCVJET-UQTMIEBXSA-N
PDB links: 6 PDB IDs match this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2-5A-dependent ribonuclease (Mus musculus) | BDBM50025002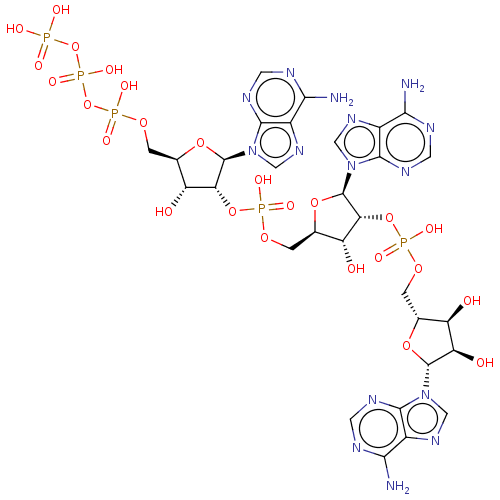 (CHEMBL404038 | ppp'A2'p5'A2'p5'A | ppp5'A2'p5'A2'p...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL KEGG PC cid PC sid PDB UniChem Similars | PubMed | n/a | n/a | 2.30 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested for activation of RNase L by measuring concentration required for 50% inhibition of protein synthesis in mouse L cell extracts | J Med Chem 29: 1015-22 (1986) BindingDB Entry DOI: 10.7270/Q2SX6C6K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||