

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: CCc1cc2[n-]c(=[OH+])c(=O)[nH]c2c(c1Cl)[N+]([O-])=O
InChI Key: InChIKey=ZVWACYCGAITBFL-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate receptor ionotropic, NMDA 1 (RAT) | BDBM50056620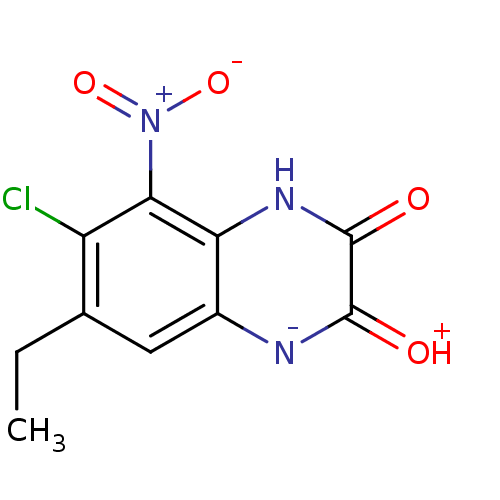 (6-Chloro-7-ethyl-5-nitro-1,4-dihydro-quinoxaline-2...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | 29 | n/a | n/a | n/a | n/a | n/a | n/a |
CoCensys Inc. Curated by ChEMBL | Assay Description Potency for the N-methyl-D-aspartate glutamate receptor 1 glycine site by displacement of [3H]5,7-dichlorokynurenic acid (DCKA) binding in rat brain ... | J Med Chem 40: 730-8 (1997) Article DOI: 10.1021/jm960654b BindingDB Entry DOI: 10.7270/Q2NZ86RJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||