

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: OC(=O)C1CCc2c(C1)[nH]c1ccc(Cl)cc21
InChI Key:
PDB links: 1 PDB ID matches this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta sliding clamp (Escherichia coli (strain K12)) | BDBM50096641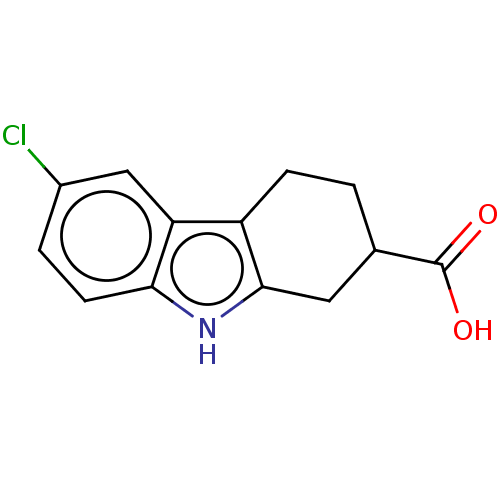 (CHEMBL3277053) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 1.66E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The University of Wollongong and The Illawarra Health and Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assay | J Med Chem 58: 4693-702 (2015) Article DOI: 10.1021/acs.jmedchem.5b00232 BindingDB Entry DOI: 10.7270/Q2KK9DJ9 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Beta sliding clamp (Escherichia coli (strain K12)) | BDBM50096641 (CHEMBL3277053) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | 2.99E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
The University of Wollongong and The Illawarra Health and Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA sliding clamp protein using N-fluorescein (FAM)-QLDLF-OH tracer by fluorescence polarization assay | J Med Chem 58: 4693-702 (2015) Article DOI: 10.1021/acs.jmedchem.5b00232 BindingDB Entry DOI: 10.7270/Q2KK9DJ9 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||