

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM50383300 CHEMBL2029610
SMILES: [#6]\[#6]-1=[#6](-[#6])/[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-c2ccc3ccccc3c2)-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H]1-[#6]-c1ccc(-[#8])cc1
InChI Key: InChIKey=QRXBSCHWFCNZSX-MXRRXXMQSA-N
Data: 1 IC50
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C-X-C chemokine receptor type 4 (CXCR4) (Homo sapiens (Human)) | BDBM50383300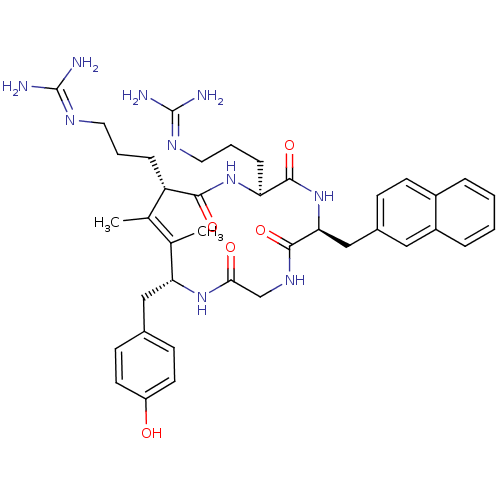 (CHEMBL2029610) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.50E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Kyoto University Curated by ChEMBL | Assay Description Displacement of [125I]SDF-1alpha from CXCR4 expressed in HEK293 cell membrane after 1 hr | J Med Chem 55: 2746-57 (2012) Article DOI: 10.1021/jm2016914 BindingDB Entry DOI: 10.7270/Q2FJ2HTW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||