

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM50495465 CHEMBL3108817
SMILES: [H][C@@]12C[C@]1([H])N([C@@H](C2)C(=O)N[C@@H]1[C@@H](Cc2ccccc12)OCC#CC#CCO[C@@H]1Cc2ccccc2[C@@H]1NC(=O)[C@@H]1C[C@]2([H])C[C@]2([H])N1C(=O)[C@@H](NC(=O)[C@H](C)NC)C1CCCCC1)C(=O)[C@@H](NC(=O)[C@H](C)NC)C1CCCCC1
InChI Key: InChIKey=TXZDEORYKQDMGZ-UCCYDSEHSA-N
Data: 1 EC50
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baculoviral IAP repeat-containing protein 2 (Homo sapiens (Human)) | BDBM50495465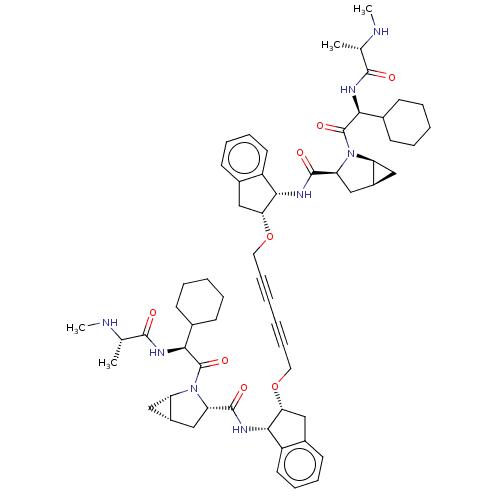 (CHEMBL3108817) | PDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | n/a | 0.100 | n/a | n/a | n/a | n/a |
AstraZeneca Curated by ChEMBL | Assay Description Binding affinity to cIAP1 in human MDA-MB-231 cells assessed as induction of protein degradation after 1 hr by ELISA | J Med Chem 56: 9897-919 (2013) Article DOI: 10.1021/jm401075x BindingDB Entry DOI: 10.7270/Q23T9M6C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||