

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM613 (2R,4S)-2-[(R)-(ethylcarbamoyl)(3-phenylpropanamido)methyl]-N-(2-{[(2R,4S)-2-[(R)-(ethylcarbamoyl)(3-phenylpropanamido)methyl]-5,5-dimethyl-1,3-thiazolidin-4-yl]formamido}ethyl)-5,5-dimethyl-1,3-thiazolidine-4-carboxamide::Penicillin Et(NH)2 Sym dimer ::penicillin deriv. 26
SMILES: [H][C@]1(N[C@@H](C(=O)NCCNC(=O)[C@@H]2N[C@]([H])(SC2(C)C)[C@H](NC(=O)CCc2ccccc2)C(=O)NCC)C(C)(C)S1)[C@H](NC(=O)CCc1ccccc1)C(=O)NCC
InChI Key: InChIKey=OSVARXBLQYUJJF-GDWNOHHZSA-N
Data: 1 IC50
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 Protease (Human immunodeficiency virus type 1) | BDBM613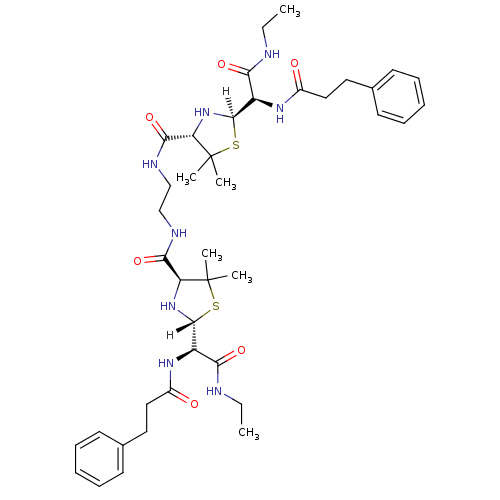 ((2R,4S)-2-[(R)-(ethylcarbamoyl)(3-phenylpropanamid...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.40 | n/a | n/a | n/a | n/a | 6.0 | 37 |
Glaxo Group Research Ltd. | Assay Description IC50 values were obtained by assaying the enzyme against the synthetic substrate peptide KQGTVSFNFPQIT, which was tritiated at the proline residue, a... | J Med Chem 36: 3120-8 (1993) Article DOI: 10.1021/jm00073a011 BindingDB Entry DOI: 10.7270/Q24F1NXP | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||