

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM781 4-hydroxy-3-(1-phenylethyl)-2H,5H,6H,7H,8H,9H,10H-cycloocta[b]pyran-2-one::Cyclooctylpyranone 6
SMILES: CC(c1ccccc1)c1c(O)c2CCCCCCc2oc1=O
InChI Key: InChIKey=NGTPYSMBKIFYEF-UHFFFAOYSA-N
Data: 1 Other
PDB links: 3 PDB IDs contain this monomer as substructures. 3 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 Protease (Human immunodeficiency virus type 1) | BDBM781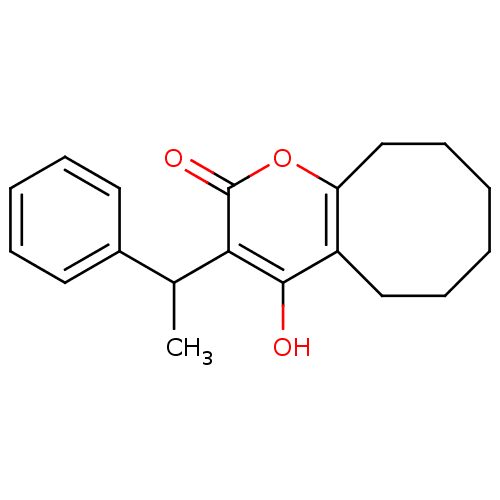 (4-hydroxy-3-(1-phenylethyl)-2H,5H,6H,7H,8H,9H,10H-...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 5.0 | 22 |
Upjohn | Assay Description HIV-1 protease was purified and refolded from E. coli inclusion bodies. The substrate used spans the p17-p24 processing site (R-V-S-Q-N-Y-P-I-V-Q-N-K... | J Med Chem 38: 1884-91 (1995) Article DOI: 10.1021/jm00011a008 BindingDB Entry DOI: 10.7270/Q2Q81B80 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||