 Found 31 hits with Last Name = 'batey' and Initial = 'ra'
Found 31 hits with Last Name = 'batey' and Initial = 'ra' Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589806

(CHEMBL5177469)Show SMILES O=C(CNCc1cc2ccccc2[nH]1)N[C@H]1CC[C@H](CC1)c1nc2ccccc2[nH]1 |r,wU:15.16,18.23,(-1.11,.83,;-1.88,2.16,;-3.42,2.16,;-4.19,.83,;-5.73,.83,;-6.5,-.51,;-6.03,-1.97,;-7.27,-2.88,;-7.43,-4.4,;-8.84,-5.03,;-10.08,-4.13,;-9.93,-2.6,;-8.52,-1.97,;-8.04,-.51,;-1.11,3.49,;.43,3.49,;1.2,4.83,;2.74,4.83,;3.51,3.49,;2.74,2.16,;1.2,2.16,;5.05,3.49,;5.95,2.25,;7.42,2.72,;8.76,1.95,;10.08,2.73,;10.08,4.26,;8.75,5.03,;7.42,4.26,;5.95,4.74,)| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | 4.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589802

(CHEMBL5177515)Show SMILES CCCCNCC(=O)N[C@H](Cc1ccc(cc1)C(C)(C)C)C(=O)N[C@@H](Cc1cccc(OC)c1)C(=O)NC |r| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | 4.70E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589804

(CHEMBL5191076)Show SMILES CNC(=O)[C@H](Cc1ccc(C)cc1)NC(=O)[C@@H](C\C=C\c1ccccc1)NC(=O)CNCc1ccccc1 |r| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | 4.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589801
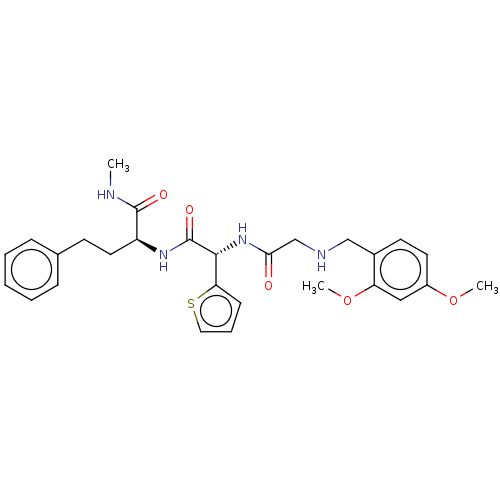
(CHEMBL5208789)Show SMILES CNC(=O)[C@H](CCc1ccccc1)NC(=O)[C@H](NC(=O)CNCc1ccc(OC)cc1OC)c1cccs1 |r| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | 5.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589797
(CHEMBL5189044) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | 1.89E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589800

(CHEMBL5197897)Show SMILES CC(C)C[C@H](NC(=O)CNC(=O)[C@@H]1CCCN1)C(=O)N[C@@H](Cc1c[nH]c2ccccc12)C(O)=O |r| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 2.14E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589799
(CHEMBL5175764) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 3.55E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589791

(CHEMBL5179578) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 4.93E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589790

(CHEMBL5194502) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 5.50E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589796

(CHEMBL5193368) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 6.48E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50017043
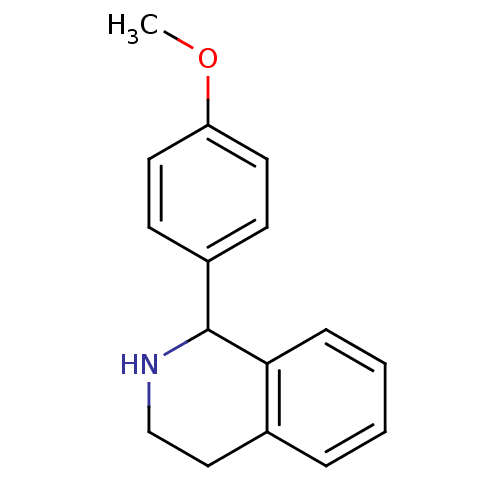
(1-(4-Methoxy-phenyl)-1,2,3,4-tetrahydro-isoquinoli...)Show InChI InChI=1S/C16H17NO/c1-18-14-8-6-13(7-9-14)16-15-5-3-2-4-12(15)10-11-17-16/h2-9,16-17H,10-11H2,1H3 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 8.98E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589787

(CHEMBL5179037) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 9.08E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589788
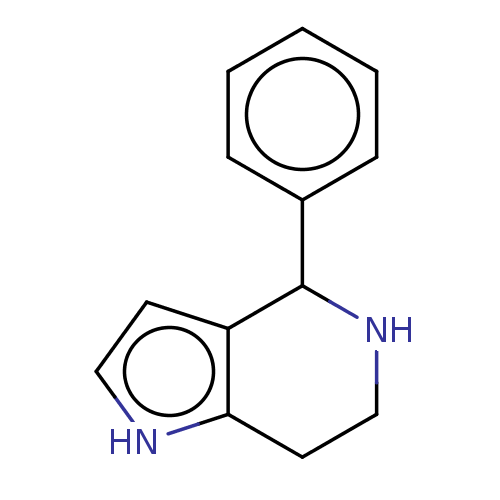
(CHEMBL5188512) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 1.08E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589785
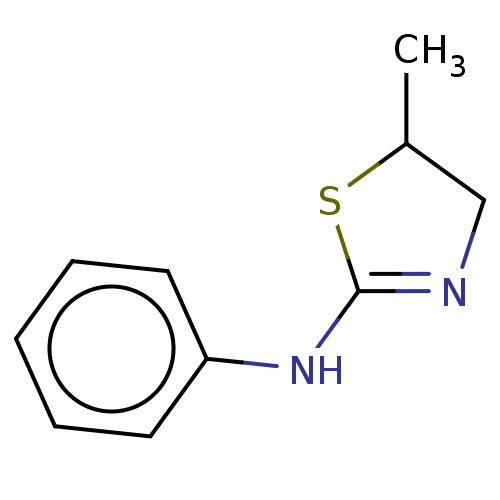
(CHEMBL1600521) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 1.13E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589784
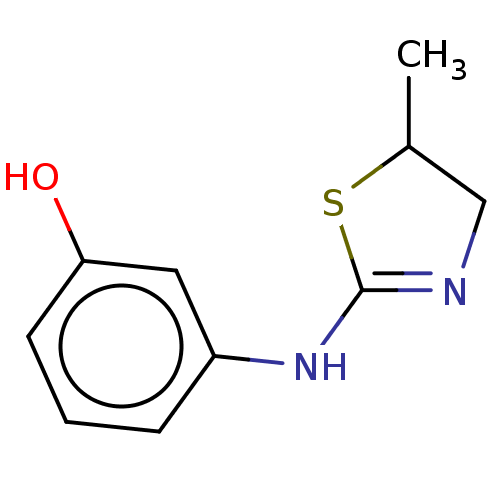
(CHEMBL5186109) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | 1.49E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589795

(CHEMBL5195783) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 1.49E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589805

(CHEMBL5191415)Show SMILES CNC(=O)C1N(Cc2ccccc12)C(=O)[C@@H](Cc1c(C)[nH]c2ccccc12)NC(=O)CNCCOc1ccc(C)cc1 |r| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 1.88E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589803

(CHEMBL5208155)Show SMILES CNC(=O)[C@H](CC1CCCCC1)NC(=O)[C@@H](NC(=O)[C@H]1NC[C@@H]1c1ccccc1)c1ccsc1 |r| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 1.88E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589793
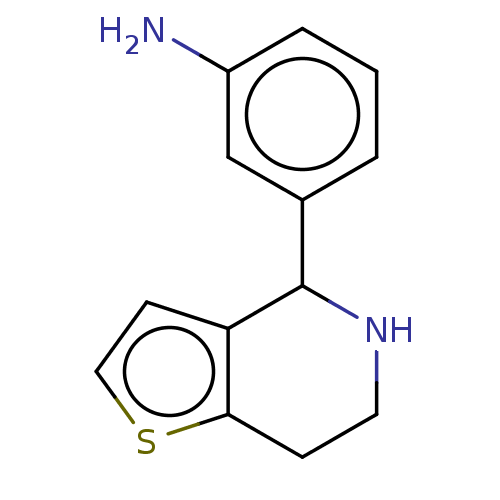
(CHEMBL5198959) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 2.35E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589798
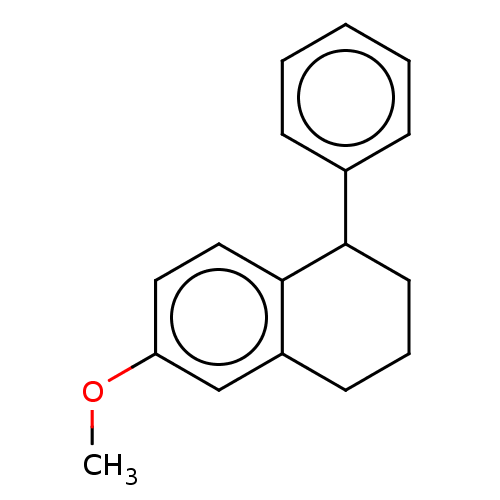
(CHEMBL5181147) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | >2.50E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589794

(CHEMBL5203207) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | >2.50E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589789

(CHEMBL5192951) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | >2.50E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589786
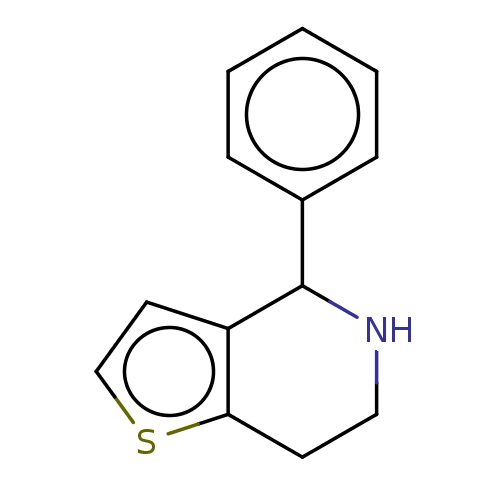
(CHEMBL2179396) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | >2.50E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589792
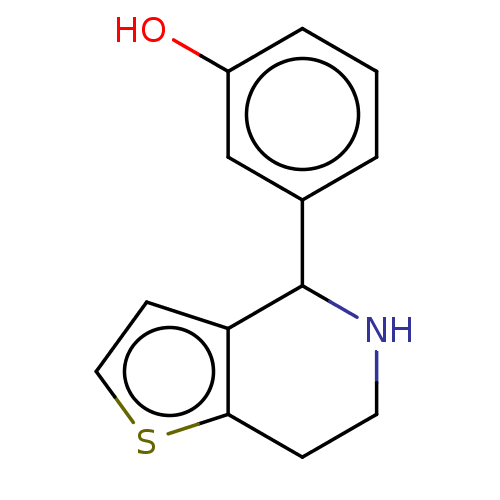
(CHEMBL5205965) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | >2.50E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589783

(CHEMBL1998455) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 2.64E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589784

(CHEMBL5186109) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 1.10E+5 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589797

(CHEMBL5189044) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 17 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589782
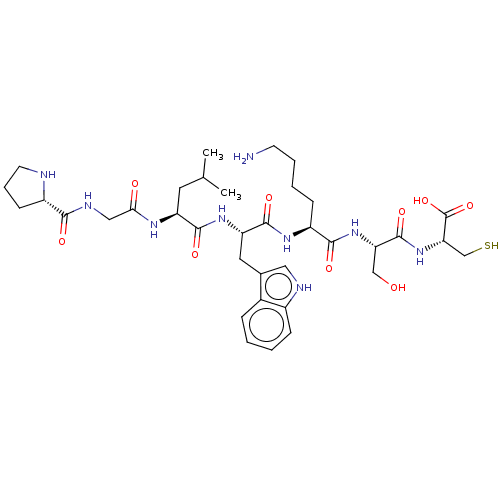
(CHEMBL5195622)Show SMILES CC(C)C[C@H](NC(=O)CNC(=O)[C@@H]1CCCN1)C(=O)N[C@@H](Cc1c[nH]c2ccccc12)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CO)C(=O)N[C@@H](CS)C(O)=O |r| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 1.90E+3 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | |
Glucose-induced degradation protein 4 homolog
(Homo sapiens) | BDBM50589806

(CHEMBL5177469)Show SMILES O=C(CNCc1cc2ccccc2[nH]1)N[C@H]1CC[C@H](CC1)c1nc2ccccc2[nH]1 |r,wU:15.16,18.23,(-1.11,.83,;-1.88,2.16,;-3.42,2.16,;-4.19,.83,;-5.73,.83,;-6.5,-.51,;-6.03,-1.97,;-7.27,-2.88,;-7.43,-4.4,;-8.84,-5.03,;-10.08,-4.13,;-9.93,-2.6,;-8.52,-1.97,;-8.04,-.51,;-1.11,3.49,;.43,3.49,;1.2,4.83,;2.74,4.83,;3.51,3.49,;2.74,2.16,;1.2,2.16,;5.05,3.49,;5.95,2.25,;7.42,2.72,;8.76,1.95,;10.08,2.73,;10.08,4.26,;8.75,5.03,;7.42,4.26,;5.95,4.74,)| | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 80 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00509
BindingDB Entry DOI: 10.7270/Q2JD51RW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent Clp protease proteolytic subunit
(Escherichia coli (strain K12)) | BDBM50028480
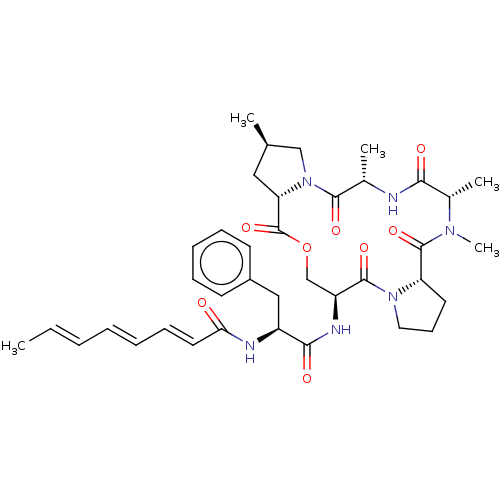
(CHEMBL3342324)Show SMILES [H][C@@]12CCCN1C(=O)[C@H](COC(=O)[C@]1([H])C[C@@H](C)CN1C(=O)[C@H](C)NC(=O)[C@H](C)N(C)C2=O)NC(=O)[C@H](Cc1ccccc1)NC(=O)\C=C\C=C\C=C\C |r| Show InChI InChI=1S/C38H50N6O8/c1-6-7-8-9-13-18-32(45)40-28(21-27-15-11-10-12-16-27)34(47)41-29-23-52-38(51)31-20-24(2)22-44(31)35(48)25(3)39-33(46)26(4)42(5)37(50)30-17-14-19-43(30)36(29)49/h6-13,15-16,18,24-26,28-31H,14,17,19-23H2,1-5H3,(H,39,46)(H,40,45)(H,41,47)/b7-6+,9-8+,18-13+/t24-,25+,26+,28+,29+,30+,31+/m1/s1 | PDB
KEGG
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | n/a | 300 | n/a | n/a | n/a | n/a | n/a |
University of Toronto
Curated by ChEMBL
| Assay Description
Activation of Escherichia coli ClpP by ITC method |
J Nat Prod 77: 2170-81 (2014)
Article DOI: 10.1021/np500158q
BindingDB Entry DOI: 10.7270/Q22R3T71 |
More data for this
Ligand-Target Pair | |
ATP-dependent Clp protease proteolytic subunit
(Escherichia coli (strain K12)) | BDBM50028481
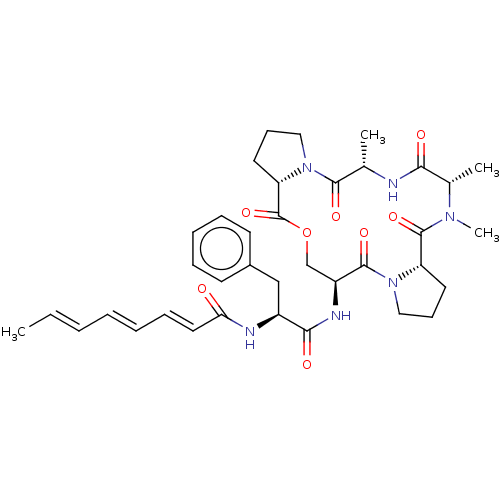
(CHEMBL3342325)Show SMILES [H][C@@]12CCCN1C(=O)[C@H](C)NC(=O)[C@H](C)N(C)C(=O)[C@]1([H])CCCN1C(=O)[C@H](COC2=O)NC(=O)[C@H](Cc1ccccc1)NC(=O)\C=C\C=C\C=C\C |r| Show InChI InChI=1S/C37H48N6O8/c1-5-6-7-8-12-19-31(44)39-27(22-26-15-10-9-11-16-26)33(46)40-28-23-51-37(50)30-18-14-21-43(30)34(47)24(2)38-32(45)25(3)41(4)36(49)29-17-13-20-42(29)35(28)48/h5-12,15-16,19,24-25,27-30H,13-14,17-18,20-23H2,1-4H3,(H,38,45)(H,39,44)(H,40,46)/b6-5+,8-7+,19-12+/t24-,25-,27-,28-,29-,30-/m0/s1 | PDB
KEGG
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | n/a | 700 | n/a | n/a | n/a | n/a | n/a |
University of Toronto
Curated by ChEMBL
| Assay Description
Activation of Escherichia coli ClpP by ITC method |
J Nat Prod 77: 2170-81 (2014)
Article DOI: 10.1021/np500158q
BindingDB Entry DOI: 10.7270/Q22R3T71 |
More data for this
Ligand-Target Pair | |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data