Ceramide glucosyltransferase
(Homo sapiens (Human)) | BDBM576435
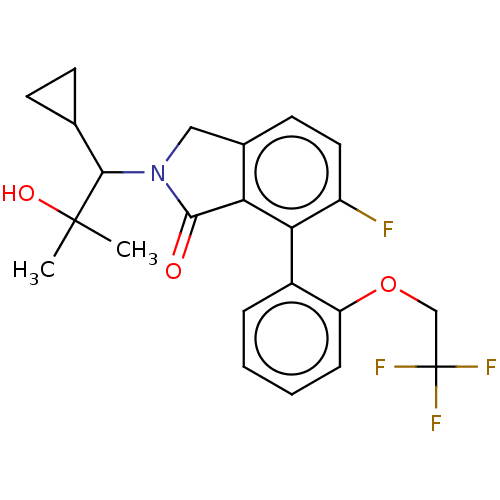
(WO2022115301, Example 21)Show SMILES CC(C)(O)C(C1CC1)N1Cc2ccc(F)c(c2C1=O)-c1ccccc1OCC(F)(F)F |(2.99,-.96,;3.76,-2.29,;5.3,-2.29,;4.53,-3.63,;2.99,-3.63,;3.76,-4.96,;3.76,-6.5,;5.1,-5.73,;1.45,-3.63,;.55,-4.87,;-.92,-4.4,;-2.25,-5.17,;-3.58,-4.4,;-3.58,-2.85,;-4.92,-2.08,;-2.25,-2.08,;-.92,-2.85,;.55,-2.38,;1.02,-.91,;-2.25,-.54,;-.92,.23,;-.92,1.77,;-2.25,2.54,;-3.58,1.77,;-3.58,.23,;-4.92,-.54,;-6.25,.23,;-7.58,-.54,;-8.92,.23,;-7.58,-2.08,;-6.25,-1.31,)| | NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| WIPO WO2022115301
| n/a | n/a | n/a | n/a | 0.910 | n/a | n/a | n/a | n/a |