 Found 2 hits in this display
Found 2 hits in this display Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Muscarinic acetylcholine receptor M2
(Homo sapiens (Human)) | BDBM280442
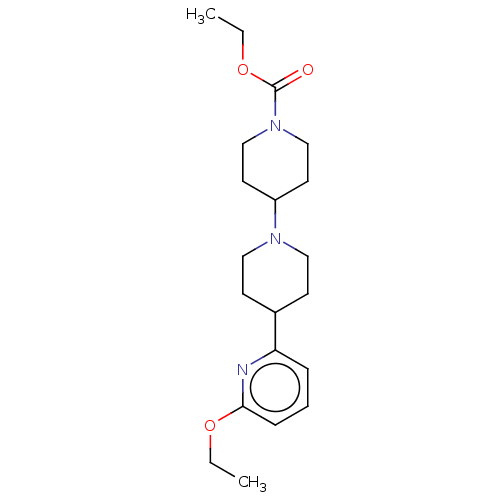
(US10030012, Example 72 | ethyl 4-[4-(6- ethoxypyri...)Show InChI InChI=1S/C20H31N3O3/c1-3-25-19-7-5-6-18(21-19)16-8-12-22(13-9-16)17-10-14-23(15-11-17)20(24)26-4-2/h5-7,16-17H,3-4,8-15H2,1-2H3 | PDB
UniProtKB/SwissProt
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| US Patent
| n/a | n/a | n/a | n/a | 2.02E+3 | n/a | n/a | n/a | n/a |
Heptares Therapeutics Limited
US Patent
| |
US Patent US10030012 (2018)
BindingDB Entry DOI: 10.7270/Q2KK9DTW |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M4
(Homo sapiens (Human)) | BDBM280442

(US10030012, Example 72 | ethyl 4-[4-(6- ethoxypyri...)Show InChI InChI=1S/C20H31N3O3/c1-3-25-19-7-5-6-18(21-19)16-8-12-22(13-9-16)17-10-14-23(15-11-17)20(24)26-4-2/h5-7,16-17H,3-4,8-15H2,1-2H3 | PDB
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| US Patent
| n/a | n/a | n/a | n/a | 270 | n/a | n/a | n/a | n/a |
Heptares Therapeutics Limited
US Patent
| Assay Description
The functional activity of compounds at the M4 and M2 receptors was determined by measuring changes in the level of intracellular calcium ions caused... |
US Patent US10030012 (2018)
BindingDB Entry DOI: 10.7270/Q2KK9DTW |
More data for this
Ligand-Target Pair | |