 Found 4 hits of ic50 for monomerid = 15374
Found 4 hits of ic50 for monomerid = 15374 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Heat shock protein HSP 90-beta
(Homo sapiens (Human)) | BDBM15374
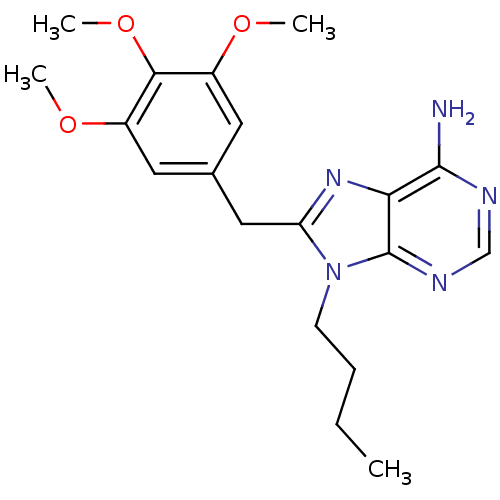
(9-butyl-8-(3,4,5-trimethoxybenzyl)-9H-purin-6-amin...)Show SMILES CCCCn1c(Cc2cc(OC)c(OC)c(OC)c2)nc2c(N)ncnc12 Show InChI InChI=1S/C19H25N5O3/c1-5-6-7-24-15(23-16-18(20)21-11-22-19(16)24)10-12-8-13(25-2)17(27-4)14(9-12)26-3/h8-9,11H,5-7,10H2,1-4H3,(H2,20,21,22) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| DrugBank
MMDB
PDB
Article
PubMed
| n/a | n/a | 1.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Memorial Sloan-Kettering Cancer Center
Curated by ChEMBL
| Assay Description
Inhibition of Hsp90 in human MCF7 cells assessed as Her2 degradation |
Bioorg Med Chem 17: 2225-35 (2009)
Article DOI: 10.1016/j.bmc.2008.10.087
BindingDB Entry DOI: 10.7270/Q2T72JCS |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Receptor tyrosine-protein kinase erbB-2
(Homo sapiens (Human)) | BDBM15374

(9-butyl-8-(3,4,5-trimethoxybenzyl)-9H-purin-6-amin...)Show SMILES CCCCn1c(Cc2cc(OC)c(OC)c(OC)c2)nc2c(N)ncnc12 Show InChI InChI=1S/C19H25N5O3/c1-5-6-7-24-15(23-16-18(20)21-11-22-19(16)24)10-12-8-13(25-2)17(27-4)14(9-12)26-3/h8-9,11H,5-7,10H2,1-4H3,(H2,20,21,22) | PDB
MMDB
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 4.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1016/j.ejmech.2022.114516
BindingDB Entry DOI: 10.7270/Q2TQ65N5 |
More data for this
Ligand-Target Pair | |
Heat shock protein HSP 90-alpha/90-beta
(Homo sapiens (Human)) | BDBM15374

(9-butyl-8-(3,4,5-trimethoxybenzyl)-9H-purin-6-amin...)Show SMILES CCCCn1c(Cc2cc(OC)c(OC)c(OC)c2)nc2c(N)ncnc12 Show InChI InChI=1S/C19H25N5O3/c1-5-6-7-24-15(23-16-18(20)21-11-22-19(16)24)10-12-8-13(25-2)17(27-4)14(9-12)26-3/h8-9,11H,5-7,10H2,1-4H3,(H2,20,21,22) | PDB
Reactome pathway
KEGG
UniProtKB/SwissProt
antibodypedia
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
PubMed
| n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
RiboTargets Ltd
Curated by ChEMBL
| Assay Description
Inhibition of ATP-ase activity in human colon tumour cell line (HCT116) |
Bioorg Med Chem Lett 14: 325-8 (2003)
BindingDB Entry DOI: 10.7270/Q2P55MX0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM15374

(9-butyl-8-(3,4,5-trimethoxybenzyl)-9H-purin-6-amin...)Show SMILES CCCCn1c(Cc2cc(OC)c(OC)c(OC)c2)nc2c(N)ncnc12 Show InChI InChI=1S/C19H25N5O3/c1-5-6-7-24-15(23-16-18(20)21-11-22-19(16)24)10-12-8-13(25-2)17(27-4)14(9-12)26-3/h8-9,11H,5-7,10H2,1-4H3,(H2,20,21,22) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd
| Assay Description
The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... |
Chem Biol 11: 775-85 (2004)
Article DOI: 10.1016/j.chembiol.2004.03.033
BindingDB Entry DOI: 10.7270/Q2RX99BC |
More data for this
Ligand-Target Pair | |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data