

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D(1A) dopamine receptor (Homo sapiens (Human)) | BDBM60917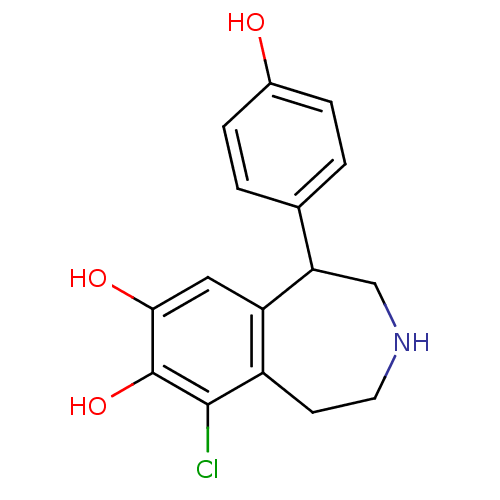 (9-chloranyl-5-(4-hydroxyphenyl)-2,3,4,5-tetrahydro...) | PDB KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Similars | DrugBank PDB Article PubMed | n/a | n/a | n/a | 28 | n/a | n/a | n/a | n/a | n/a |
VA Medical Center Curated by ChEMBL | Assay Description Equilibrium dissociation constant against recombinant Dopamine receptor D1A expressed in COS7 cells | J Med Chem 39: 850-9 (1996) Article DOI: 10.1021/jm950447w BindingDB Entry DOI: 10.7270/Q2XG9RT3 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D(2) dopamine receptor (Rattus norvegicus (rat)) | BDBM60917 (9-chloranyl-5-(4-hydroxyphenyl)-2,3,4,5-tetrahydro...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Similars | PubMed | n/a | n/a | n/a | 1.30E+3 | n/a | n/a | n/a | n/a | n/a |
University of Texas Curated by ChEMBL | Assay Description In vitro affinity at wild type Dopamine receptor D2 on C6 (glioma) cell membranes. | J Med Chem 43: 3005-19 (2000) BindingDB Entry DOI: 10.7270/Q24J0FS2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(2) dopamine receptor (Rattus norvegicus (rat)) | BDBM60917 (9-chloranyl-5-(4-hydroxyphenyl)-2,3,4,5-tetrahydro...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Similars | PubMed | n/a | n/a | n/a | 1.90E+3 | n/a | n/a | n/a | n/a | n/a |
University of Texas Curated by ChEMBL | Assay Description In vitro affinity at mutant D2 receptor (S197A) in C6 (glioma) cell membranes. | J Med Chem 43: 3005-19 (2000) BindingDB Entry DOI: 10.7270/Q24J0FS2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(2) dopamine receptor (Rattus norvegicus (rat)) | BDBM60917 (9-chloranyl-5-(4-hydroxyphenyl)-2,3,4,5-tetrahydro...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Similars | PubMed | n/a | n/a | n/a | 2.70E+3 | n/a | n/a | n/a | n/a | n/a |
University of Texas Curated by ChEMBL | Assay Description In vitro affinity at mutant D2 receptor (S194A) in C6 (glioma) cell membranes. | J Med Chem 43: 3005-19 (2000) BindingDB Entry DOI: 10.7270/Q24J0FS2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(2) dopamine receptor (Rattus norvegicus (rat)) | BDBM60917 (9-chloranyl-5-(4-hydroxyphenyl)-2,3,4,5-tetrahydro...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Similars | PubMed | n/a | n/a | n/a | 1.30E+4 | n/a | n/a | n/a | n/a | n/a |
University of Texas Curated by ChEMBL | Assay Description In vitro affinity at mutant D2 receptor (S194A) in C6 (glioma) cell membranes. | J Med Chem 43: 3005-19 (2000) BindingDB Entry DOI: 10.7270/Q24J0FS2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||