

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome polyprotein (Hepatitis C virus (HCV genotype 1a, isolate H)) | BDBM12268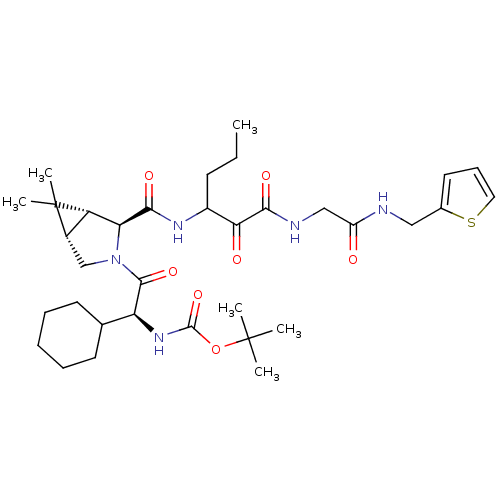 (1,1-Dimethylethyl-[1(S)-cyclohexyl-2-[(1R,5S)-6,6-...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 60 | -41.9 | n/a | n/a | n/a | n/a | n/a | 6.5 | 30 |
Schering-Plough Research Institute | Assay Description Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I... | J Med Chem 49: 6074-86 (2006) Article DOI: 10.1021/jm060325b BindingDB Entry DOI: 10.7270/Q2DF6PF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||