

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (Rattus norvegicus (rat)) | BDBM14755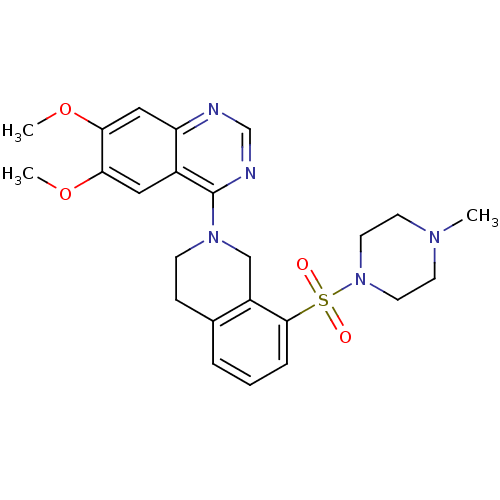 (6,7-dimethoxy-4-[8-(4-methylpiperazin-1-yl)sulfony...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents | MMDB PDB Article PubMed | 25 | -43.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 22 |
Pfizer | Assay Description PDE activity was monitored by measuring the hydrolysis of [3H]-cAMP to [3H]-AMP using a scintillation proximity assay (SPA). [3H]-AMP was captured by... | J Med Chem 50: 182-5 (2007) Article DOI: 10.1021/jm060653b BindingDB Entry DOI: 10.7270/Q2930RDX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||