

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16240 (2-(carboxymethyl)-1-oxo-1,2-dihydronaphtho[1,2-d]i...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 140 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16242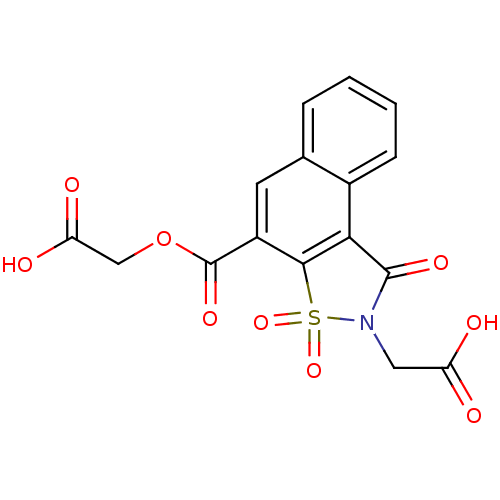 (2-{[4-(carboxymethyl)-3,5,5-trioxo-5-thia-4-azatri...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 550 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16243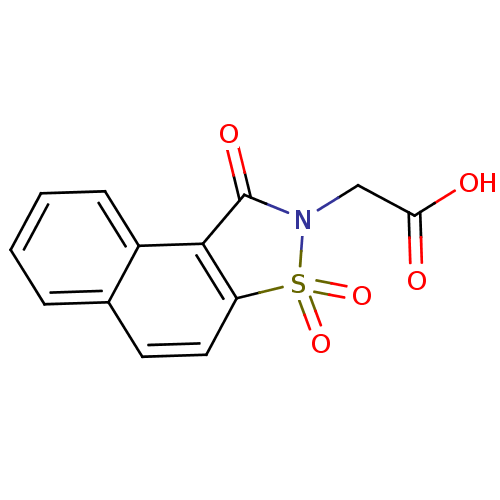 (2-(3,5,5-trioxo-5-thia-4-azatricyclo[7.4.0.0^{2,6}...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.00E+4 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16248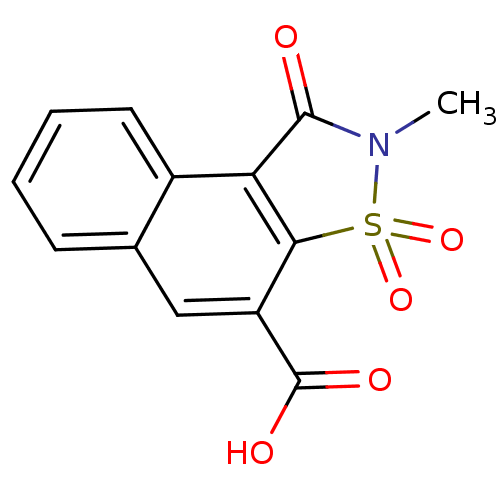 (4-methyl-3,5,5-trioxo-5-thia-4-azatricyclo[7.4.0.0...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 8.80E+4 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16247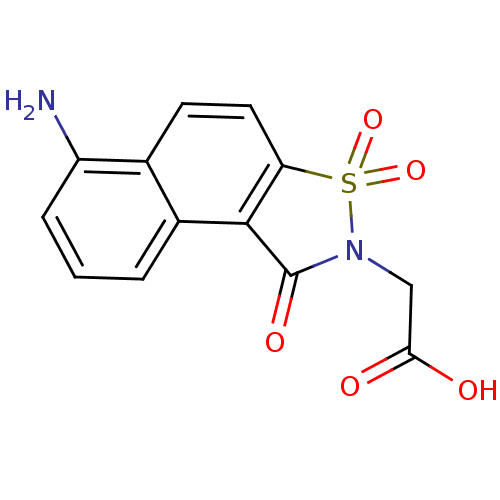 (2-(10-amino-3,5,5-trioxo-5-thia-4-azatricyclo[7.4....) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.00E+5 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16246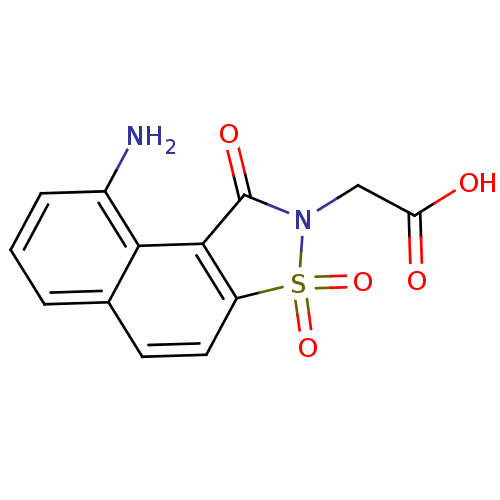 (2-(13-amino-3,5,5-trioxo-5-thia-4-azatricyclo[7.4....) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.00E+5 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16245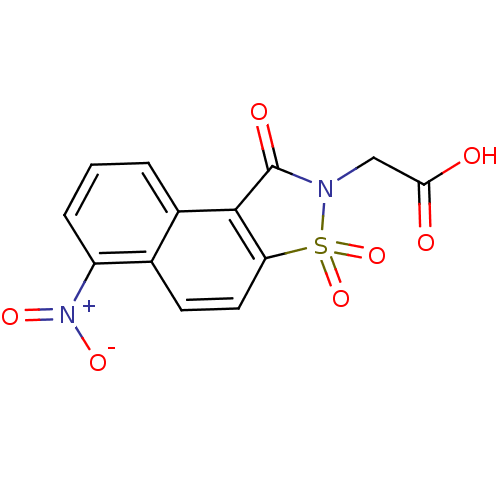 (2-(10-nitro-3,5,5-trioxo-5-thia-4-azatricyclo[7.4....) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.80E+5 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16244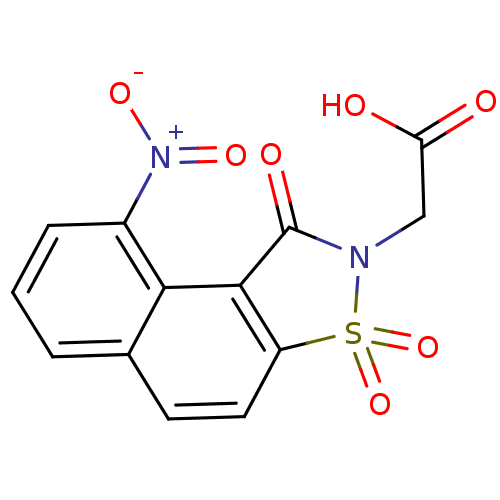 (2-(13-nitro-3,5,5-trioxo-5-thia-4-azatricyclo[7.4....) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.90E+5 | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Aldo-keto reductase family 1 member B1 (Homo sapiens (Human)) | BDBM16249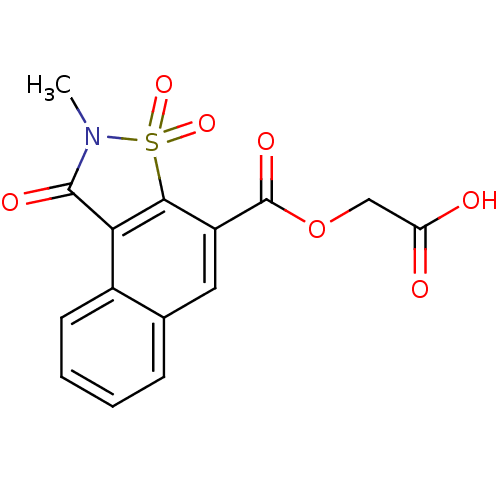 (2-[(4-methyl-3,5,5-trioxo-5-thia-4-azatricyclo[7.4...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 6.2 | 30 |
University of Marburg | Assay Description The activity of the test enzyme was determined spectrophotometrically by monitoring the change in absorbance at 340 nm, which is due to the oxidation... | J Mol Biol 369: 186-97 (2007) Article DOI: 10.1016/j.jmb.2007.03.021 BindingDB Entry DOI: 10.7270/Q2BK19MR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||