 Found 7 hits of kd data for polymerid = 50002756,50002926,50003406
Found 7 hits of kd data for polymerid = 50002756,50002926,50003406 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
3-dehydroquinate dehydratase
(Helicobacter pylori) | BDBM14672
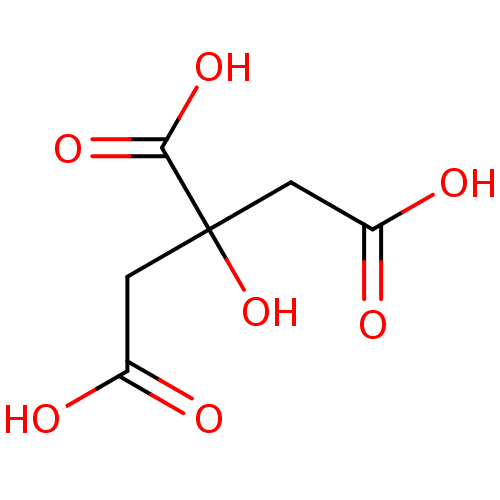
(2-hydroxypropane-1,2,3-tricarboxylic acid | CHEMBL...)Show InChI InChI=1S/C6H8O7/c7-3(8)1-6(13,5(11)12)2-4(9)10/h13H,1-2H2,(H,7,8)(H,9,10)(H,11,12) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
KEGG
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 2.50E+3 | n/a | n/a | n/a | n/a | n/a |
University of Glasgow
Curated by ChEMBL
| Assay Description
Binding affinity for Helicobacter pylori DHQase 2 |
J Med Chem 49: 1282-90 (2006)
Article DOI: 10.1021/jm0505361
BindingDB Entry DOI: 10.7270/Q27P8XZF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
3-dehydroquinate dehydratase
(Streptomyces coelicolor) | BDBM14672

(2-hydroxypropane-1,2,3-tricarboxylic acid | CHEMBL...)Show InChI InChI=1S/C6H8O7/c7-3(8)1-6(13,5(11)12)2-4(9)10/h13H,1-2H2,(H,7,8)(H,9,10)(H,11,12) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
KEGG
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | n/a | 7.30E+3 | n/a | n/a | n/a | n/a | n/a |
University of Glasgow
Curated by ChEMBL
| Assay Description
Binding affinity for Streptomyces coelicolor DHQase 2 |
J Med Chem 49: 1282-90 (2006)
Article DOI: 10.1021/jm0505361
BindingDB Entry DOI: 10.7270/Q27P8XZF |
More data for this
Ligand-Target Pair | |
3-dehydroquinate dehydratase
(Helicobacter pylori) | BDBM50182477
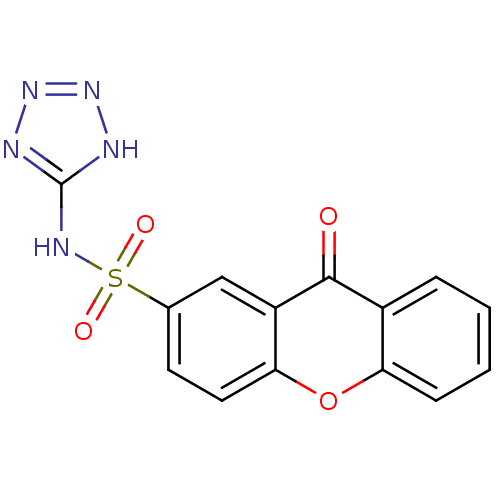
(CHEMBL202311 | N-TETRAZOL-5-YL 9-OXO-9H-XANTHENE-2...)Show SMILES O=c1c2ccccc2oc2ccc(cc12)S(=O)(=O)Nc1nnn[nH]1 Show InChI InChI=1S/C14H9N5O4S/c20-13-9-3-1-2-4-11(9)23-12-6-5-8(7-10(12)13)24(21,22)17-14-15-18-19-16-14/h1-7H,(H2,15,16,17,18,19) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
UniChem
| MMDB
Article
PubMed
| n/a | n/a | n/a | 2.00E+4 | n/a | n/a | n/a | n/a | n/a |
University of Glasgow
Curated by ChEMBL
| Assay Description
Binding affinity for Helicobacter pylori DHQase 2 |
J Med Chem 49: 1282-90 (2006)
Article DOI: 10.1021/jm0505361
BindingDB Entry DOI: 10.7270/Q27P8XZF |
More data for this
Ligand-Target Pair | |
3-dehydroquinate dehydratase
(Streptomyces coelicolor) | BDBM50170812

((1R,4R,5R)-1,4,5-Trihydroxy-cyclohex-2-enecarboxyl...)Show InChI InChI=1S/C7H10O5/c8-4-1-2-7(12,6(10)11)3-5(4)9/h1-2,4-5,8-9,12H,3H2,(H,10,11)/t4-,5-,7+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 3.00E+4 | n/a | n/a | n/a | n/a | n/a |
University of Glasgow
Curated by ChEMBL
| Assay Description
Binding affinity for Streptomyces coelicolor DHQase 2 |
J Med Chem 49: 1282-90 (2006)
Article DOI: 10.1021/jm0505361
BindingDB Entry DOI: 10.7270/Q27P8XZF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
3-dehydroquinate dehydratase
(Mycobacterium tuberculosis) | BDBM50170812

((1R,4R,5R)-1,4,5-Trihydroxy-cyclohex-2-enecarboxyl...)Show InChI InChI=1S/C7H10O5/c8-4-1-2-7(12,6(10)11)3-5(4)9/h1-2,4-5,8-9,12H,3H2,(H,10,11)/t4-,5-,7+/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 2.00E+5 | n/a | n/a | n/a | n/a | n/a |
University of Glasgow
Curated by ChEMBL
| Assay Description
Binding affinity for Mycobacterium tuberculosis DHQase 2 |
J Med Chem 49: 1282-90 (2006)
Article DOI: 10.1021/jm0505361
BindingDB Entry DOI: 10.7270/Q27P8XZF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
3-dehydroquinate dehydratase
(Streptomyces coelicolor) | BDBM50182477

(CHEMBL202311 | N-TETRAZOL-5-YL 9-OXO-9H-XANTHENE-2...)Show SMILES O=c1c2ccccc2oc2ccc(cc12)S(=O)(=O)Nc1nnn[nH]1 Show InChI InChI=1S/C14H9N5O4S/c20-13-9-3-1-2-4-11(9)23-12-6-5-8(7-10(12)13)24(21,22)17-14-15-18-19-16-14/h1-7H,(H2,15,16,17,18,19) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 2.30E+5 | n/a | n/a | n/a | n/a | n/a |
University of Glasgow
Curated by ChEMBL
| Assay Description
Binding affinity for Streptomyces coelicolor DHQase 2 |
J Med Chem 49: 1282-90 (2006)
Article DOI: 10.1021/jm0505361
BindingDB Entry DOI: 10.7270/Q27P8XZF |
More data for this
Ligand-Target Pair | |
3-dehydroquinate dehydratase
(Helicobacter pylori) | BDBM50170812

((1R,4R,5R)-1,4,5-Trihydroxy-cyclohex-2-enecarboxyl...)Show InChI InChI=1S/C7H10O5/c8-4-1-2-7(12,6(10)11)3-5(4)9/h1-2,4-5,8-9,12H,3H2,(H,10,11)/t4-,5-,7+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 3.70E+5 | n/a | n/a | n/a | n/a | n/a |
University of Glasgow
Curated by ChEMBL
| Assay Description
Binding affinity for Helicobacter pylori DHQase 2 |
J Med Chem 49: 1282-90 (2006)
Article DOI: 10.1021/jm0505361
BindingDB Entry DOI: 10.7270/Q27P8XZF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data