

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dimer of Gag-Pol polyprotein [489-587] (98/99 = 99%)† (Human immunodeficiency virus type 1) | BDBM851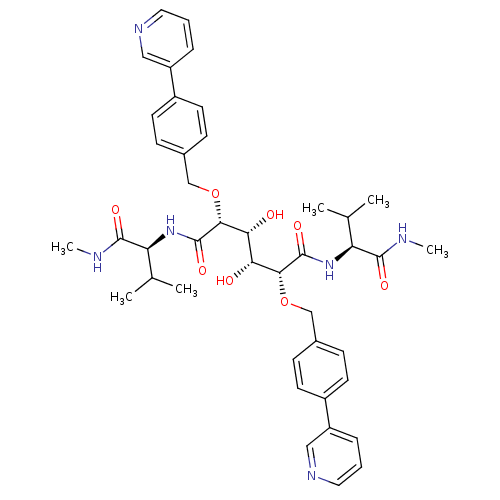 ((2R,3R,4R,5R)-3,4-dihydroxy-N,N'-bis[(1S)-2-methyl...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.100 | -58.0 | n/a | n/a | n/a | n/a | n/a | 5.0 | 30 |
Uppsala University | Assay Description A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid... | Eur J Biochem 270: 1746-58 (2003) Article DOI: 10.1046/j.1432-1033.2003.03533.x BindingDB Entry DOI: 10.7270/Q2NZ85WX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599] (99/99 = 100%)† (Human immunodeficiency virus type 1) | BDBM851 ((2R,3R,4R,5R)-3,4-dihydroxy-N,N'-bis[(1S)-2-methyl...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 0.300 | -55.3 | n/a | n/a | n/a | n/a | n/a | 5.0 | 30 |
Uppsala University | Assay Description Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a... | J Med Chem 42: 3835-44 (1999) Article DOI: 10.1021/jm9910371 BindingDB Entry DOI: 10.7270/Q2T151VX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||