

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L-lactate dehydrogenase B chain (333/333 = 100%)† (Homo sapiens (Human)) | BDBM23222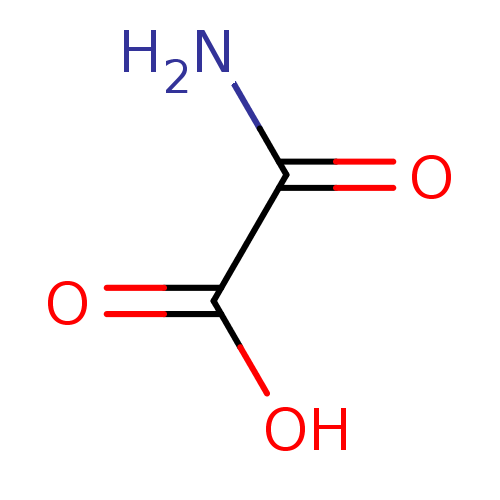 (Oxalamic acid | Oxamate | Oxamate, 3 | Oxamidic Ac...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents | DrugBank PDB Article PubMed | 9.44E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
National Cancer Institute-CRO Curated by ChEMBL | Assay Description Competitive inhibition of human LDH1 in presence of NADH | J Med Chem 59: 487-96 (2016) Article DOI: 10.1021/acs.jmedchem.5b00168 BindingDB Entry DOI: 10.7270/Q2B27X41 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| L-lactate dehydrogenase B chain (333/333 = 100%)† (Homo sapiens (Human)) | BDBM23222 (Oxalamic acid | Oxamate | Oxamate, 3 | Oxamidic Ac...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents | DrugBank PDB Article PubMed | n/a | n/a | 3.38E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Genentech Curated by ChEMBL | Assay Description Inhibition of human recombinant carboxy-terminal His-tagged LDHB by UV endpoint assay | Bioorg Med Chem Lett 24: 3764-71 (2014) Article DOI: 10.1016/j.bmcl.2014.06.076 BindingDB Entry DOI: 10.7270/Q2445P46 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| L-lactate dehydrogenase B chain (333/333 = 100%)† (Homo sapiens (Human)) | BDBM23222 (Oxalamic acid | Oxamate | Oxamate, 3 | Oxamidic Ac...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents | DrugBank PDB Article PubMed | n/a | n/a | 3.38E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Genentech Curated by ChEMBL | Assay Description Inhibition of human recombinant carboxy-terminal his-tagged LDHB (1 to 333) expressed in insect cells using pyruvate as substrate after 10 mins by UV... | Bioorg Med Chem Lett 23: 3186-94 (2013) Article DOI: 10.1016/j.bmcl.2013.04.001 BindingDB Entry DOI: 10.7270/Q29C6ZTT | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| L-lactate dehydrogenase B chain (326/333 = 98%)† (Mus musculus (Mouse)) | BDBM23222 (Oxalamic acid | Oxamate | Oxamate, 3 | Oxamidic Ac...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | 6.00E+4 | n/a | n/a | n/a | n/a | 7.4 | n/a |
Instituto Politécnico Nacional | Assay Description Lactate dehydrogenase activity was determined by recording the absorbance change at 340 nm produced by the oxidation of NADH. Assays were performed a... | J Enzyme Inhib Med Chem 26: 579-86 (2011) Article DOI: 10.3109/14756366.2011.566221 BindingDB Entry DOI: 10.7270/Q2R78D36 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| L-lactate dehydrogenase B chain (327/333 = 98%)† (Bos taurus (bovine)) | BDBM23222 (Oxalamic acid | Oxamate | Oxamate, 3 | Oxamidic Ac...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | n/a | n/a | 1.23E+5 | n/a | n/a | n/a | n/a | 7.0 | 25 |
University of Mississippi | Assay Description LDH was assayed spectrophotometrically for reduction of pyruvate using NADH by recording the changed in absorbance at 340 nm. The reaction was initia... | J Med Chem 50: 3841-50 (2007) Article DOI: 10.1021/jm070336k BindingDB Entry DOI: 10.7270/Q2HH6HCV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||