

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Androgen receptor (249/249 = 100%)† (Rattus norvegicus (Rat)) | BDBM18665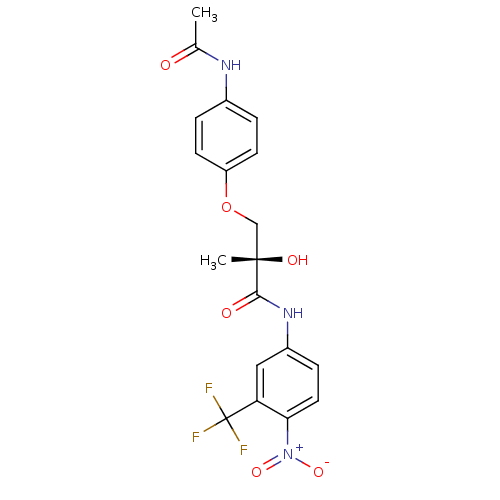 ((2S)-3-(4-acetamidophenoxy)-2-hydroxy-2-methyl-N-[...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 3.98 | -44.6 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
University of Tennessee at Memphis | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | J Med Chem 47: 993-8 (2004) Article DOI: 10.1021/jm030336u BindingDB Entry DOI: 10.7270/Q2JH3JFB | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (249/249 = 100%)† (Homo sapiens (Human)) | BDBM18665 ((2S)-3-(4-acetamidophenoxy)-2-hydroxy-2-methyl-N-[...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GTx, Inc. Curated by ChEMBL | Assay Description Agonist activity at androgen receptor (unknown origin) | J Med Chem 52: 3597-617 (2009) Article DOI: 10.1021/jm900280m BindingDB Entry DOI: 10.7270/Q2GH9HWD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (249/249 = 100%)† (Rattus norvegicus (Rat)) | BDBM18665 ((2S)-3-(4-acetamidophenoxy)-2-hydroxy-2-methyl-N-[...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 4 | -44.6 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||