

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Endolysin [L99A] (155/164 = 95%)† (Enterobacteria phage T4) | BDBM50008567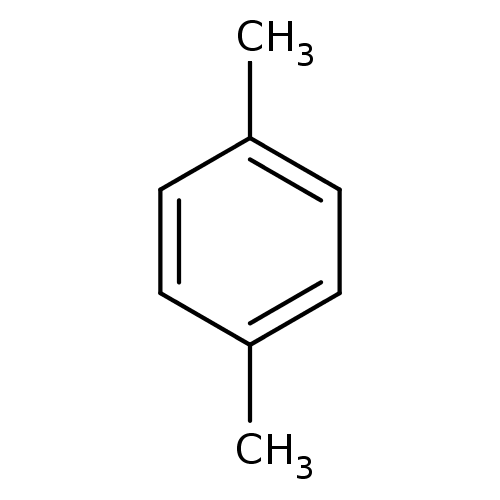 (1,4-Dimethylbenzol | 1,4-xylene | 4-methyltoluene ...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | n/a | 4.22E+5 | n/a | n/a | n/a | n/a | n/a |
University of California San Francisco Curated by ChEMBL | Assay Description Dissociation constant against T4 lysozyme mutant L99A | J Med Chem 48: 3714-28 (2005) Article DOI: 10.1021/jm0491187 BindingDB Entry DOI: 10.7270/Q2VT1SW6 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||