

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peroxisome proliferator-activated receptor gamma (282/283 > 99%)† (Homo sapiens (Human)) | BDBM50299109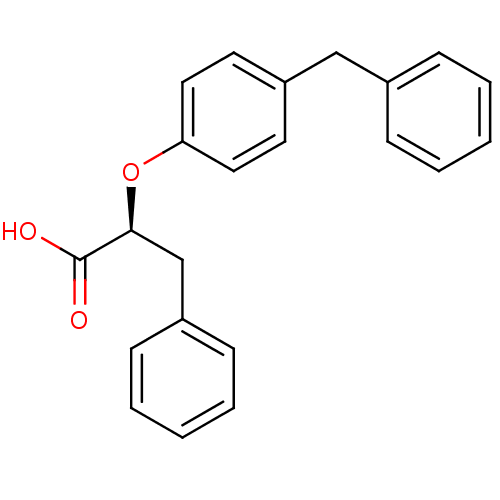 ((2S)-2-(4-benzylphenoxy)-3-phenylpropanoic acid | ...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | n/a | 580 | n/a | n/a | n/a | n/a |
Universit£ degli Studi di Bari Curated by ChEMBL | Assay Description Agonist activity at GAL4-tagged human PPARgamma ligand binding domain expressed in human HepG2 cells assessed as receptor transactivation by lucifera... | J Med Chem 52: 6382-93 (2009) Article DOI: 10.1021/jm900941b BindingDB Entry DOI: 10.7270/Q2862GHF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Peroxisome proliferator-activated receptor gamma (282/283 > 99%)† (Homo sapiens (Human)) | BDBM50299109 ((2S)-2-(4-benzylphenoxy)-3-phenylpropanoic acid | ...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | n/a | 575 | n/a | n/a | n/a | n/a |
Universit£ degli studi di Bari 'Aldo Moro' Curated by ChEMBL | Assay Description Agonist activity at GAL4-fused PPARgamma (unknown origin) expressed in human HepG2 cells by transactivation assay | Eur J Med Chem 63: 321-32 (2013) Article DOI: 10.1016/j.ejmech.2013.02.015 BindingDB Entry DOI: 10.7270/Q29G5QRP | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Peroxisome proliferator-activated receptor gamma (282/283 > 99%)† (Homo sapiens (Human)) | BDBM50299109 ((2S)-2-(4-benzylphenoxy)-3-phenylpropanoic acid | ...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | n/a | 600 | n/a | n/a | n/a | n/a |
Universit£ degli studi di Bari 'Aldo Moro' Curated by ChEMBL | Assay Description Agonist activity at GAL4-fused PPARgamma (unknown origin) expressed in human HepG2 cells by transactivation assay | Eur J Med Chem 63: 321-32 (2013) Article DOI: 10.1016/j.ejmech.2013.02.015 BindingDB Entry DOI: 10.7270/Q29G5QRP | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Peroxisome proliferator-activated receptor gamma (282/283 > 99%)† (Homo sapiens (Human)) | BDBM50299109 ((2S)-2-(4-benzylphenoxy)-3-phenylpropanoic acid | ...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | n/a | 572 | n/a | n/a | n/a | n/a |
Universit£ degli Studi di Bari Curated by ChEMBL | Assay Description Agonist activity at GAL4-tagged human PPARgamma ligand binding domain expressed in human HepG2 cells assessed as receptor transactivation by lucifera... | J Med Chem 52: 6382-93 (2009) Article DOI: 10.1021/jm900941b BindingDB Entry DOI: 10.7270/Q2862GHF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||