

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dihydrofolate reductase (157/157 = 100%)† (Staphylococcus aureus) | BDBM50329610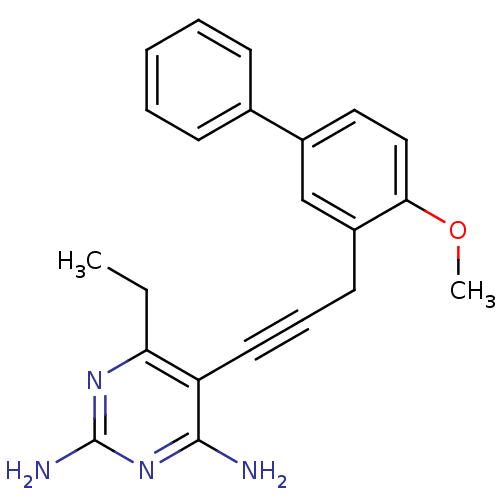 (6-ethyl-5-(3-(4-methoxybiphenyl-3-yl)prop-1-ynyl)p...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | PDB US Patent | n/a | n/a | 2.40 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Connecticut US Patent | Assay Description Heterocyclic analogs of propargyl-linked inhibitors of the third generation analogs were evaluated in enzyme inhibition assays, assessed for S. aur... | US Patent US8853228 (2014) BindingDB Entry DOI: 10.7270/Q2TD9W1T | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||