

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Histone-lysine N-methyltransferase EZH2 | ||
| Ligand | BDBM574925 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ZH2 biochemical assay (IC50) | ||
| IC50 | 0.5±n/a nM | ||
| Citation |  Brenneman, JB; Côté, A; Gehling, VS; Khanna, A; Levell, JR; Moine, L Modulators of methyl modifying enzymes, compositions and uses thereof US Patent US11459315 Publication Date 10/4/2022 Brenneman, JB; Côté, A; Gehling, VS; Khanna, A; Levell, JR; Moine, L Modulators of methyl modifying enzymes, compositions and uses thereof US Patent US11459315 Publication Date 10/4/2022 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Histone-lysine N-methyltransferase EZH2 | |||
| Name: | Histone-lysine N-methyltransferase EZH2 | ||
| Synonyms: | ENX-1 | EZH2 | EZH2_HUMAN | Enhancer of zeste homolog 2 (EZH2) | Histone-lysine N-methyltransferase EZH2 | KMT6 | Lysine N-methyltransferase 6 | ||
| Type: | Protein | ||
| Mol. Mass.: | 85367.84 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q15910 | ||
| Residue: | 746 | ||
| Sequence: |
| ||
| BDBM574925 | |||
| n/a | |||
| Name | BDBM574925 | ||
| Synonyms: | 1-((R)-1-(4-(2-methoxyethoxy)cyclohex-yl)ethyl)-2-methyl-N-((6-methyl-4-(methylthio)-2-oxo-1,2-dihydropyridin-3-yl)methyl)-1H-indole-3-carboxamide | US11459315, Example 67 (isomer 2) | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H39N3O4S | ||
| Mol. Mass. | 525.703 | ||
| SMILES | COCCOC1CC[C@@H](CC1)[C@@H](C)n1c(C)c(C(=O)NCc2c(SC)cc(C)[nH]c2=O)c2ccccc12 |wU:11.12,wD:8.8,(7.86,-4.14,;6.37,-3.74,;5.28,-4.83,;3.79,-4.43,;2.7,-5.52,;1.22,-5.12,;.13,-6.21,;-1.36,-5.81,;-1.76,-4.32,;-.67,-3.23,;.82,-3.63,;-3.25,-3.93,;-4.34,-5.01,;-3.72,-2.46,;-2.82,-1.21,;-1.28,-1.21,;-3.72,.03,;-3.25,1.5,;-4.28,2.64,;-1.74,1.82,;-1.27,3.28,;.24,3.6,;.72,5.07,;-.31,6.21,;-1.82,5.89,;2.22,5.39,;3.25,4.24,;4.76,4.56,;2.78,2.78,;1.27,2.46,;.8,.99,;-5.19,-.44,;-6.52,.33,;-7.86,-.44,;-7.86,-1.98,;-6.52,-2.75,;-5.19,-1.98,)| | ||
| Structure | 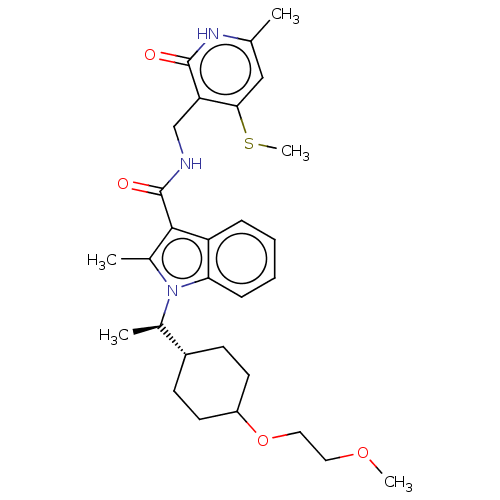
| ||