| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Urease subunit alpha [F36L]/beta/gamma |
|---|
| Ligand | BDBM24966 |
|---|
| Substrate/Competitor | BDBM24961 |
|---|
| Meas. Tech. | Enzyme Inhibition Assay |
|---|
| pH | 7±n/a |
|---|
| Temperature | 303.15±n/a K |
|---|
| Ki | 41000±2000 nM |
|---|
| IC50 | 342000±23000 nM |
|---|
| Citation |  Vassiliou, S; Grabowiecka, A; Kosikowska, P; Yiotakis, A; Kafarski, P; Berlicki, L Design, synthesis, and evaluation of novel organophosphorus inhibitors of bacterial ureases. J Med Chem51:5736-44 (2008) [PubMed] Article Vassiliou, S; Grabowiecka, A; Kosikowska, P; Yiotakis, A; Kafarski, P; Berlicki, L Design, synthesis, and evaluation of novel organophosphorus inhibitors of bacterial ureases. J Med Chem51:5736-44 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Urease subunit alpha [F36L]/beta/gamma |
|---|
| Name: | Urease subunit alpha [F36L]/beta/gamma |
|---|
| Synonyms: | Urea Amidohydrolase | Urea Amidohydrolase-pv | Urease |
|---|
| Type: | Heterotrimer of ureA, ureB and ureC subunits. Three heterotrimers associate to form the active enzyme. |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 3 components. |
|---|
| Component 1 |
| Name: | Urease subunit alpha [F36L] |
|---|
| Synonyms: | URE1_PROMH | Urea Amidohydrolase Subunit alpha | ureC |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 60882.75 |
|---|
| Organism: | Proteus vulgaris |
|---|
| Description: | P17086[F36L] |
|---|
| Residue: | 567 |
|---|
| Sequence: | MKTISRQAYADMFGPTTGDRLRLADTELFLEIEKDLTTYGEEVKFGGGKVIRDGMGQSQV
VSAECVDVLITNAIILDYWGIVKADIGIKDGRIVGIGKAGNPDVQPNVDIVIGPGTEVVA
GEGKIVTAGGIDTHIHFICPQQAQEGLVSGVTTFIGGGTGPVAGTNATTVTPGIWNMYRM
LEAVDELPINVGLFGKGCVSQPEAIREQITAGAIGLKIHEDWGATPMAIHNCLNVADEMD
VQVAIHSDTLNEGGFYEETVKAIAGRVIHVFHTEGAGGGHAPDVIKSVGEPNILPASTNP
TMPYTINTVDEHLDMLMVCHHLDPSIPEDVAFAESRIRRETIAAEDILHDMGAISVMSSD
SQAMGRVGEVILRTWQCAHKMKLQRGTLAGDSADNDNNRIKRYIAKYTINPALAHGIAHT
VGSIEKGKLADIVLWDPAFFGVKPALIIKGGMVAYAPMGDINAAIPTPQPVHYRPMYACL
GKAKYQTSMIFMSKAGIEAGVPEKLGLKSLIGRVEGCRHITKASMIHNNYVPHIELDPQT
YIVKADGVPLVCEPATELPMAQRYFLF
|
|
|
|---|
| Component 2 |
| Name: | Urease subunit beta |
|---|
| Synonyms: | UreB | Urea Amidohydrolase Subunit beta |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 12029.52 |
|---|
| Organism: | Proteus vulgaris |
|---|
| Description: | n/a |
|---|
| Residue: | 108 |
|---|
| Sequence: | MIPGEISVNQALGDIELNAGRETKIIQVANHGDRPVQVGSHYHFYEVNEALNFERESTLG
FRLNIPAGMAVRFEPGQSRTVELVAFAGKREIYGFHGKVMGKLESEKK
|
|
|
|---|
| Component 3 |
| Name: | Urease subunit gamma |
|---|
| Synonyms: | UreA | Urea Amidohydrolase Subunit gamma |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 10908.21 |
|---|
| Organism: | Proteus vulgaris |
|---|
| Description: | n/a |
|---|
| Residue: | 100 |
|---|
| Sequence: | MELTPREKDKLLLFTAGLVAERRLAKGLKLNYPEAVALISCAIMEGAREGKTVAQLMSEG
RSVLAAEQVMEGVPEMIKDIQVECTFPDGTKLVSIHDPIV
|
|
|
|---|
| BDBM24966 |
|---|
| BDBM24961 |
|---|
| Name | BDBM24966 |
|---|
| Synonyms: | organophosphorus derivative, 7 | {[(2S)-2-amino-3-hydroxypropanamido]methyl}(methyl)phosphinic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C5H13N2O4P |
|---|
| Mol. Mass. | 196.1415 |
|---|
| SMILES | CP(O)(=O)CNC(=O)[C@@H](N)CO |r| |
|---|
| Structure | 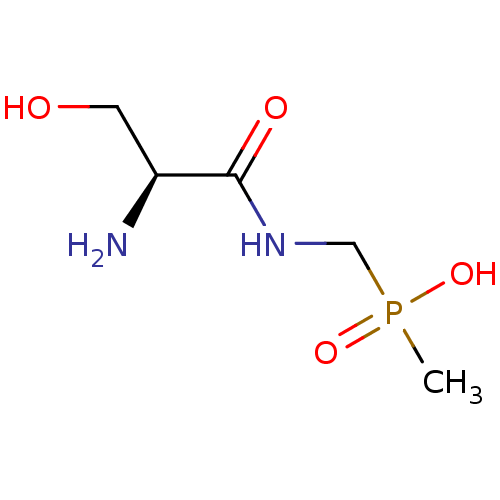
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Vassiliou, S; Grabowiecka, A; Kosikowska, P; Yiotakis, A; Kafarski, P; Berlicki, L Design, synthesis, and evaluation of novel organophosphorus inhibitors of bacterial ureases. J Med Chem51:5736-44 (2008) [PubMed] Article
Vassiliou, S; Grabowiecka, A; Kosikowska, P; Yiotakis, A; Kafarski, P; Berlicki, L Design, synthesis, and evaluation of novel organophosphorus inhibitors of bacterial ureases. J Med Chem51:5736-44 (2008) [PubMed] Article