| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM19149 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1700232 (CHEMBL4051214) |
|---|
| IC50 | 261±n/a nM |
|---|
| Citation |  Ahmad, M; Aga, MA; Bhat, JA; Kumar, B; Rouf, A; Capalash, N; Mintoo, MJ; Kumar, A; Mahajan, P; Mondhe, DM; Nargotra, A; Sharma, PR; Zargar, MA; Vishwakarma, RA; Shah, BA; Taneja, SC; Hamid, A Exploring Derivatives of Quinazoline Alkaloid l-Vasicine as Cap Groups in the Design and Biological Mechanistic Evaluation of Novel Antitumor Histone Deacetylase Inhibitors. J Med Chem60:3484-3497 (2017) [PubMed] Article Ahmad, M; Aga, MA; Bhat, JA; Kumar, B; Rouf, A; Capalash, N; Mintoo, MJ; Kumar, A; Mahajan, P; Mondhe, DM; Nargotra, A; Sharma, PR; Zargar, MA; Vishwakarma, RA; Shah, BA; Taneja, SC; Hamid, A Exploring Derivatives of Quinazoline Alkaloid l-Vasicine as Cap Groups in the Design and Biological Mechanistic Evaluation of Novel Antitumor Histone Deacetylase Inhibitors. J Med Chem60:3484-3497 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM19149 |
|---|
| n/a |
|---|
| Name | BDBM19149 |
|---|
| Synonyms: | CHEMBL98 | N-hydroxy-N'-phenyloctanediamide | SAHA | US10011611, SAHA | US10188756, Compound SAHA | US11207431, SAHA | US11505523, Compound SAHA | US9115116, SAHA | US9353061, SAHA | US9428447, SAHA | US9695181, Vorinostat | Vorinostat | Zolinza | cid_5311 | suberoylanilide hydroxamic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H20N2O3 |
|---|
| Mol. Mass. | 264.3202 |
|---|
| SMILES | ONC(=O)CCCCCCC(=O)Nc1ccccc1 |
|---|
| Structure | 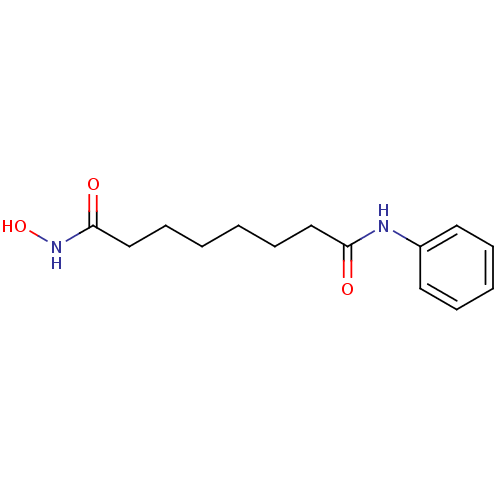
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ahmad, M; Aga, MA; Bhat, JA; Kumar, B; Rouf, A; Capalash, N; Mintoo, MJ; Kumar, A; Mahajan, P; Mondhe, DM; Nargotra, A; Sharma, PR; Zargar, MA; Vishwakarma, RA; Shah, BA; Taneja, SC; Hamid, A Exploring Derivatives of Quinazoline Alkaloid l-Vasicine as Cap Groups in the Design and Biological Mechanistic Evaluation of Novel Antitumor Histone Deacetylase Inhibitors. J Med Chem60:3484-3497 (2017) [PubMed] Article
Ahmad, M; Aga, MA; Bhat, JA; Kumar, B; Rouf, A; Capalash, N; Mintoo, MJ; Kumar, A; Mahajan, P; Mondhe, DM; Nargotra, A; Sharma, PR; Zargar, MA; Vishwakarma, RA; Shah, BA; Taneja, SC; Hamid, A Exploring Derivatives of Quinazoline Alkaloid l-Vasicine as Cap Groups in the Design and Biological Mechanistic Evaluation of Novel Antitumor Histone Deacetylase Inhibitors. J Med Chem60:3484-3497 (2017) [PubMed] Article