| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM50471953 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_153698 (CHEMBL756487) |
|---|
| Ki | 107±n/a nM |
|---|
| Citation |  Collins, JL; Blanchard, SG; Boswell, GE; Charifson, PS; Cobb, JE; Henke, BR; Hull-Ryde, EA; Kazmierski, WM; Lake, DH; Leesnitzer, LM; Lehmann, J; Lenhard, JM; Orband-Miller, LA; Gray-Nunez, Y; Parks, DJ; Plunkett, KD; Tong, WQ N-(2-Benzoylphenyl)-L-tyrosine PPARgamma agonists. 2. Structure-activity relationship and optimization of the phenyl alkyl ether moiety. J Med Chem41:5037-54 (1998) [PubMed] Article Collins, JL; Blanchard, SG; Boswell, GE; Charifson, PS; Cobb, JE; Henke, BR; Hull-Ryde, EA; Kazmierski, WM; Lake, DH; Leesnitzer, LM; Lehmann, J; Lenhard, JM; Orband-Miller, LA; Gray-Nunez, Y; Parks, DJ; Plunkett, KD; Tong, WQ N-(2-Benzoylphenyl)-L-tyrosine PPARgamma agonists. 2. Structure-activity relationship and optimization of the phenyl alkyl ether moiety. J Med Chem41:5037-54 (1998) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM50471953 |
|---|
| n/a |
|---|
| Name | BDBM50471953 |
|---|
| Synonyms: | CHEMBL356382 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C35H32N2O6 |
|---|
| Mol. Mass. | 576.6384 |
|---|
| SMILES | COc1ccc(cc1)-c1nc(CCOc2ccc(C[C@H](Nc3ccccc3C(=O)c3ccccc3)C(O)=O)cc2)c(C)o1 |
|---|
| Structure | 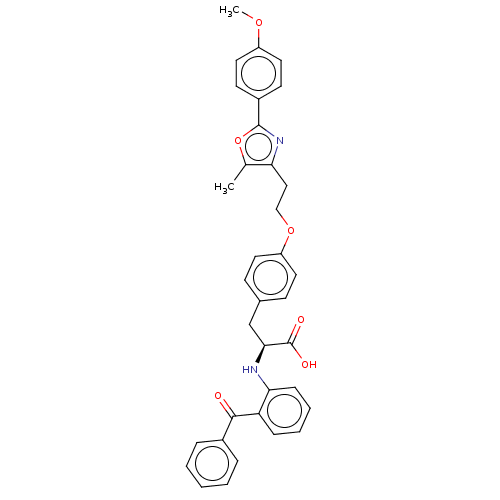
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Collins, JL; Blanchard, SG; Boswell, GE; Charifson, PS; Cobb, JE; Henke, BR; Hull-Ryde, EA; Kazmierski, WM; Lake, DH; Leesnitzer, LM; Lehmann, J; Lenhard, JM; Orband-Miller, LA; Gray-Nunez, Y; Parks, DJ; Plunkett, KD; Tong, WQ N-(2-Benzoylphenyl)-L-tyrosine PPARgamma agonists. 2. Structure-activity relationship and optimization of the phenyl alkyl ether moiety. J Med Chem41:5037-54 (1998) [PubMed] Article
Collins, JL; Blanchard, SG; Boswell, GE; Charifson, PS; Cobb, JE; Henke, BR; Hull-Ryde, EA; Kazmierski, WM; Lake, DH; Leesnitzer, LM; Lehmann, J; Lenhard, JM; Orband-Miller, LA; Gray-Nunez, Y; Parks, DJ; Plunkett, KD; Tong, WQ N-(2-Benzoylphenyl)-L-tyrosine PPARgamma agonists. 2. Structure-activity relationship and optimization of the phenyl alkyl ether moiety. J Med Chem41:5037-54 (1998) [PubMed] Article