| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4A/4B/4C/4D |
|---|
| Ligand | BDBM50473722 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_157194 (CHEMBL768607) |
|---|
| IC50 | 0.630957±n/a nM |
|---|
| Citation |  Van der Mey, M; Boss, H; Couwenberg, D; Hatzelmann, A; Sterk, GJ; Goubitz, K; Schenk, H; Timmerman, H Novel selective phosphodiesterase (PDE4) inhibitors. 4. Resolution, absolute configuration, and PDE4 inhibitory activity of cis-tetra- and cis-hexahydrophthalazinones. J Med Chem45:2526-33 (2002) [PubMed] Article Van der Mey, M; Boss, H; Couwenberg, D; Hatzelmann, A; Sterk, GJ; Goubitz, K; Schenk, H; Timmerman, H Novel selective phosphodiesterase (PDE4) inhibitors. 4. Resolution, absolute configuration, and PDE4 inhibitory activity of cis-tetra- and cis-hexahydrophthalazinones. J Med Chem45:2526-33 (2002) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 4A/4B/4C/4D |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4A/4B/4C/4D |
|---|
| Synonyms: | Phosphodiesterase 4 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 788178 |
|---|
| Components: | This complex has 4 components. |
|---|
| Component 1 |
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE2 | PDE46 | PDE4A | PDE4A_HUMAN | Phosphodiesterase 4 (PDE4) | Phosphodiesterase 4A | Phosphodiesterase 4A (PDE4) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 98113.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P27815 |
|---|
| Residue: | 886 |
|---|
| Sequence: | MEPPTVPSERSLSLSLPGPREGQATLKPPPQHLWRQPRTPIRIQQRGYSDSAERAERERQ
PHRPIERADAMDTSDRPGLRTTRMSWPSSFHGTGTGSGGAGGGSSRRFEAENGPTPSPGR
SPLDSQASPGLVLHAGAATSQRRESFLYRSDSDYDMSPKTMSRNSSVTSEAHAEDLIVTP
FAQVLASLRSVRSNFSLLTNVPVPSNKRSPLGGPTPVCKATLSEETCQQLARETLEELDW
CLEQLETMQTYRSVSEMASHKFKRMLNRELTHLSEMSRSGNQVSEYISTTFLDKQNEVEI
PSPTMKEREKQQAPRPRPSQPPPPPVPHLQPMSQITGLKKLMHSNSLNNSNIPRFGVKTD
QEELLAQELENLNKWGLNIFCVSDYAGGRSLTCIMYMIFQERDLLKKFRIPVDTMVTYML
TLEDHYHADVAYHNSLHAADVLQSTHVLLATPALDAVFTDLEILAALFAAAIHDVDHPGV
SNQFLINTNSELALMYNDESVLENHHLAVGFKLLQEDNCDIFQNLSKRQRQSLRKMVIDM
VLATDMSKHMTLLADLKTMVETKKVTSSGVLLLDNYSDRIQVLRNMVHCADLSNPTKPLE
LYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTASVEKSQVGFIDYIVHPLWETWADL
VHPDAQEILDTLEDNRDWYYSAIRQSPSPPPEEESRGPGHPPLPDKFQFELTLEEEEEEE
ISMAQIPCTAQEALTAQGLSGVEEALDATIAWEASPAQESLEVMAQEASLEAELEAVYLT
QQAQSTGSAPVAPDEFSSREEFVVAVSHSSPSALALQSPLLPAWRTLSVSEHAPGLPGLP
STAAEVEAQREHQAAKRACSACAGTFGEDTSALPAPGGGGSGGDPT
|
|
|
|---|
| Component 2 |
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4D |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE3 | PDE43 | PDE4D | PDE4D_HUMAN | Phosphodiesterase 4D |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 91092.69 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q08499 |
|---|
| Residue: | 809 |
|---|
| Sequence: | MEAEGSSAPARAGSGEGSDSAGGATLKAPKHLWRHEQHHQYPLRQPQFRLLHPHHHLPPP
PPPSPQPQPQCPLQPPPPPPLPPPPPPPGAARGRYASSGATGRVRHRGYSDTERYLYCRA
MDRTSYAVETGHRPGLKKSRMSWPSSFQGLRRFDVDNGTSAGRSPLDPMTSPGSGLILQA
NFVHSQRRESFLYRSDSDYDLSPKSMSRNSSIASDIHGDDLIVTPFAQVLASLRTVRNNF
AALTNLQDRAPSKRSPMCNQPSINKATITEEAYQKLASETLEELDWCLDQLETLQTRHSV
SEMASNKFKRMLNRELTHLSEMSRSGNQVSEFISNTFLDKQHEVEIPSPTQKEKEKKKRP
MSQISGVKKLMHSSSLTNSSIPRFGVKTEQEDVLAKELEDVNKWGLHVFRIAELSGNRPL
TVIMHTIFQERDLLKTFKIPVDTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLST
PALEAVFTDLEILAAIFASAIHDVDHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGF
KLLQEENCDIFQNLTKKQRQSLRKMVIDIVLATDMSKHMNLLADLKTMVETKKVTSSGVL
LLDNYSDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGMEISPMC
DKHNASVEKSQVGFIDYIVHPLWETWADLVHPDAQDILDTLEDNREWYQSTIPQSPSPAP
DDPEEGRQGQTEKFQFELTLEEDGESDTEKDSGSQVEEDTSCSDSKTLCTQDSESTEIPL
DEQVEEEAVGEEEESQPEACVIDDRSPDT
|
|
|
|---|
| Component 3 |
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4B |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE4 | Isoform PDE4B1 | PDE32 | PDE4B | PDE4B1 | PDE4B_HUMAN | Phosphodiesterase 4B | Phosphodiesterase 4B (PDE4B) | Phosphodiesterase 4B (PDE4B1) | Phosphodiesterase Type 4 (PDE4B) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 83318.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07343 |
|---|
| Residue: | 736 |
|---|
| Sequence: | MKKSRSVMTVMADDNVKDYFECSLSKSYSSSSNTLGIDLWRGRRCCSGNLQLPPLSQRQS
ERARTPEGDGISRPTTLPLTTLPSIAITTVSQECFDVENGPSPGRSPLDPQASSSAGLVL
HATFPGHSQRRESFLYRSDSDYDLSPKAMSRNSSLPSEQHGDDLIVTPFAQVLASLRSVR
NNFTILTNLHGTSNKRSPAASQPPVSRVNPQEESYQKLAMETLEELDWCLDQLETIQTYR
SVSEMASNKFKRMLNRELTHLSEMSRSGNQVSEYISNTFLDKQNDVEIPSPTQKDREKKK
KQQLMTQISGVKKLMHSSSLNNTSISRFGVNTENEDHLAKELEDLNKWGLNIFNVAGYSH
NRPLTCIMYAIFQERDLLKTFRISSDTFITYMMTLEDHYHSDVAYHNSLHAADVAQSTHV
LLSTPALDAVFTDLEILAAIFAAAIHDVDHPGVSNQFLINTNSELALMYNDESVLENHHL
AVGFKLLQEEHCDIFMNLTKKQRQTLRKMVIDMVLATDMSKHMSLLADLKTMVETKKVTS
SGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEI
SPMCDKHTASVEKSQVGFIDYIVHPLWETWADLVQPDAQDILDTLEDNRNWYQSMIPQSP
SPPLDEQNRDCQGLMEKFQFELTLDEEDSEGPEKEGEGHSYFSSTKTLCVIDPENRDSLG
ETDIDIATEDKSPVDT
|
|
|
|---|
| Component 4 |
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4C |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE1 | NP_000914.2 | PDE21 | PDE4C | PDE4C1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 79875.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q08493 |
|---|
| Residue: | 712 |
|---|
| Sequence: | MENLGVGEGAEACSRLSRSRGRHSMTRAPKHLWRQPRRPIRIQQRFYSDPDKSAGCRERD
LSPRPELRKSRLSWPVSSCRRFDLENGLSCGRRALDPQSSPGLGRIMQAPVPHSQRRESF
LYRSDSDYELSPKAMSRNSSVASDLHGEDMIVTPFAQVLASLRTVRSNVAALARQQCLGA
AKQGPVGNPSSSNQLPPAEDTGQKLALETLDELDWCLDQLETLQTRHSVGEMASNKFKRI
LNRELTHLSETSRSGNQVSEYISRTFLDQQTEVELPKVTAEEAPQPMSRISGLHGLCHSA
SLSSATVPRFGVQTDQEEQLAKELEDTNKWGLDVFKVAELSGNRPLTAIIFSIFQERDLL
KTFQIPADTLATYLLMLEGHYHANVAYHNSLHAADVAQSTHVLLATPALEAVFTDLEILA
ALFASAIHDVDHPGVSNQFLINTNSELALMYNDASVLENHHLAVGFKLLQAENCDIFQNL
SAKQRLSLRRMVIDMVLATDMSKHMNLLADLKTMVETKKVTSLGVLLLDNYSDRIQVLQN
LVHCADLSNPTKPLPLYRQWTDRIMAEFFQQGDRERESGLDISPMCDKHTASVEKSQVGF
IDYIAHPLWETWADLVHPDAQDLLDTLEDNREWYQSKIPRSPSDLTNPERDGPDRFQFEL
TLEEAEEEDEEEEEEGEETALAKEALELPDTELLSPEAGPDPGDLPLDNQRT
|
|
|
|---|
| BDBM50473722 |
|---|
| n/a |
|---|
| Name | BDBM50473722 |
|---|
| Synonyms: | CHEMBL315565 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H32N2O3 |
|---|
| Mol. Mass. | 420.5439 |
|---|
| SMILES | COc1ccc(cc1OC)C1=NN(C2C3CC4CC(C3)CC2C4)C(=O)C2CC=CCC12 |c:31,t:11,TLB:12:13:15:18.19.17,THB:20:21:15:18.19.17,20:18:15:13.21.22,17:16:13:18.20.19,17:18:13:16.15.22,(9.5,-12.21,;10.84,-11.45,;12.18,-12.21,;13.51,-11.45,;14.85,-12.21,;14.85,-13.76,;13.51,-14.53,;12.18,-13.76,;10.84,-14.53,;9.5,-13.76,;16.18,-14.53,;16.16,-16.07,;17.5,-16.82,;17.49,-18.35,;18.84,-19.05,;20.05,-20.33,;18.84,-21.58,;18.8,-23.08,;17.63,-21.39,;18.9,-20.58,;16.26,-20.69,;16.2,-19.16,;16.74,-20.49,;18.84,-16.07,;20.17,-16.83,;18.83,-14.53,;20.17,-13.76,;20.17,-12.23,;18.84,-11.45,;17.5,-12.21,;17.5,-13.74,)| |
|---|
| Structure | 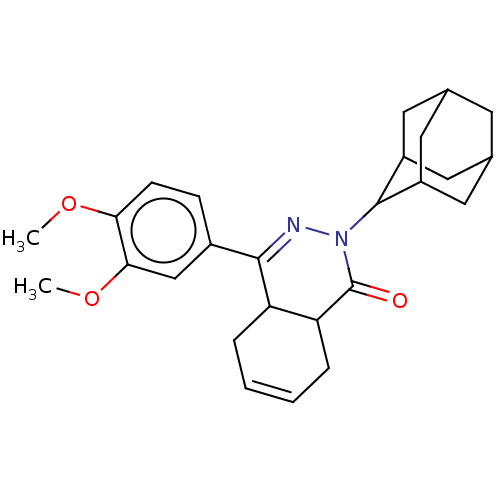
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Van der Mey, M; Boss, H; Couwenberg, D; Hatzelmann, A; Sterk, GJ; Goubitz, K; Schenk, H; Timmerman, H Novel selective phosphodiesterase (PDE4) inhibitors. 4. Resolution, absolute configuration, and PDE4 inhibitory activity of cis-tetra- and cis-hexahydrophthalazinones. J Med Chem45:2526-33 (2002) [PubMed] Article
Van der Mey, M; Boss, H; Couwenberg, D; Hatzelmann, A; Sterk, GJ; Goubitz, K; Schenk, H; Timmerman, H Novel selective phosphodiesterase (PDE4) inhibitors. 4. Resolution, absolute configuration, and PDE4 inhibitory activity of cis-tetra- and cis-hexahydrophthalazinones. J Med Chem45:2526-33 (2002) [PubMed] Article