

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | RNA-directed RNA polymerase | ||
| Ligand | BDBM50537585 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1969960 (CHEMBL4602778) | ||
| IC50 | 5.0±n/a nM | ||
| Citation |  Ganta, NM; Gedda, G; Rathnakar, B; Satyanarayana, M; Yamajala, B; Ahsan, MJ; Jadav, SS; Balaraju, T A review on HCV inhibitors: Significance of non-structural polyproteins. Eur J Med Chem164:576-601 (2019) [PubMed] Article Ganta, NM; Gedda, G; Rathnakar, B; Satyanarayana, M; Yamajala, B; Ahsan, MJ; Jadav, SS; Balaraju, T A review on HCV inhibitors: Significance of non-structural polyproteins. Eur J Med Chem164:576-601 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| RNA-directed RNA polymerase | |||
| Name: | RNA-directed RNA polymerase | ||
| Synonyms: | Hepatitis C virus NS5B RNA-dependent RNA polymerase | NS5B protein | ||
| Type: | Protein | ||
| Mol. Mass.: | 25173.95 | ||
| Organism: | Hepatitis C virus | ||
| Description: | Q8JXU8 | ||
| Residue: | 229 | ||
| Sequence: |
| ||
| BDBM50537585 | |||
| n/a | |||
| Name | BDBM50537585 | ||
| Synonyms: | CHEMBL4646449 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H21F2N3O4S | ||
| Mol. Mass. | 497.514 | ||
| SMILES | Cc1ccc2c(c(C(=O)NS(=O)(=O)C3CC3)n(Cc3cc(F)ccc3F)c2c1)-c1ccc[nH]c1=O |(25.71,-7.97,;27.04,-8.74,;28.37,-7.96,;29.71,-8.73,;29.71,-10.28,;30.86,-11.3,;30.24,-12.71,;31.01,-14.04,;32.55,-14.03,;30.25,-15.38,;31.02,-16.7,;32.35,-15.92,;32.36,-17.46,;30.26,-18.04,;28.93,-18.82,;30.27,-19.59,;28.71,-12.55,;27.92,-13.88,;26.38,-13.86,;25.64,-12.52,;24.1,-12.5,;23.34,-11.16,;23.31,-13.83,;24.07,-15.17,;25.61,-15.18,;26.37,-16.52,;28.39,-11.05,;27.04,-10.28,;32.36,-10.98,;33.39,-12.12,;34.89,-11.8,;35.37,-10.34,;34.34,-9.19,;32.82,-9.51,;31.79,-8.37,)| | ||
| Structure | 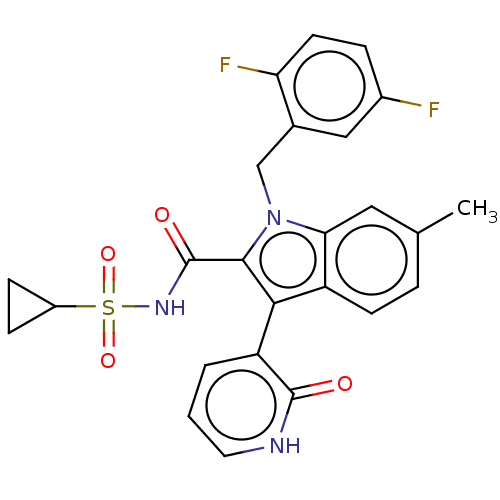
| ||