| Reaction Details |
|---|
 | Report a problem with these data |
| Target | TP53-binding protein 1 |
|---|
| Ligand | BDBM50496593 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1300217 (CHEMBL3136681) |
|---|
| IC50 | 14500±n/a nM |
|---|
| Citation |  Camerino, MA; Zhong, N; Dong, A; Dickson, BM; James, LI; Baughman, BM; Norris, JL; Kireev, DB; Janzen, WP; Arrowsmith, CH; Frye, SV The structure-activity relationships of L3MBTL3 inhibitors: flexibility of the dimer interface. Medchemcomm4:1501-1507 (2013) [PubMed] Article Camerino, MA; Zhong, N; Dong, A; Dickson, BM; James, LI; Baughman, BM; Norris, JL; Kireev, DB; Janzen, WP; Arrowsmith, CH; Frye, SV The structure-activity relationships of L3MBTL3 inhibitors: flexibility of the dimer interface. Medchemcomm4:1501-1507 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| TP53-binding protein 1 |
|---|
| Name: | TP53-binding protein 1 |
|---|
| Synonyms: | 53BP1 | Methyl-lysine binding protein (53BP1) | TP53BP1 | TP53B_HUMAN | Tumor suppressor p53-binding protein 1 | p53-binding protein 1 | p53BP1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 213442.98 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_107658 |
|---|
| Residue: | 1972 |
|---|
| Sequence: | MDPTGSQLDSDFSQQDTPCLIIEDSQPESQVLEDDSGSHFSMLSRHLPNLQTHKENPVLD
VVSNPEQTAGEERGDGNSGFNEHLKENKVADPVDSSNLDTCGSISQVIEQLPQPNRTSSV
LGMSVESAPAVEEEKGEELEQKEKEKEEDTSGNTTHSLGAEDTASSQLGFGVLELSQSQD
VEENTVPYEVDKEQLQSVTTNSGYTRLSDVDANTAIKHEEQSNEDIPIAEQSSKDIPVTA
QPSKDVHVVKEQNPPPARSEDMPFSPKASVAAMEAKEQLSAQELMESGLQIQKSPEPEVL
STQEDLFDQSNKTVSSDGCSTPSREEGGCSLASTPATTLHLLQLSGQRSLVQDSLSTNSS
DLVAPSPDAFRSTPFIVPSSPTEQEGRQDKPMDTSVLSEEGGEPFQKKLQSGEPVELENP
PLLPESTVSPQASTPISQSTPVFPPGSLPIPSQPQFSHDIFIPSPSLEEQSNDGKKDGDM
HSSSLTVECSKTSEIEPKNSPEDLGLSLTGDSCKLMLSTSEYSQSPKMESLSSHRIDEDG
ENTQIEDTEPMSPVLNSKFVPAENDSILMNPAQDGEVQLSQNDDKTKGDDTDTRDDISIL
ATGCKGREETVAEDVCIDLTCDSGSQAVPSPATRSEALSSVLDQEEAMEIKEHHPEEGSS
GSEVEEIPETPCESQGEELKEENMESVPLHLSLTETQSQGLCLQKEMPKKECSEAMEVET
SVISIDSPQKLAILDQELEHKEQEAWEEATSEDSSVVIVDVKEPSPRVDVSCEPLEGVEK
CSDSQSWEDIAPEIEPCAENRLDTKEEKSVEYEGDLKSGTAETEPVEQDSSQPSLPLVRA
DDPLRLDQELQQPQTQEKTSNSLTEDSKMANAKQLSSDAEAQKLGKPSAHASQSFCESSS
ETPFHFTLPKEGDIIPPLTGATPPLIGHLKLEPKRHSTPIGISNYPESTIATSDVMSESM
VETHDPILGSGKGDSGAAPDVDDKLCLRMKLVSPETEASEESLQFNLEKPATGERKNGST
AVAESVASPQKTMSVLSCICEARQENEARSEDPPTTPIRGNLLHFPSSQGEEEKEKLEGD
HTIRQSQQPMKPISPVKDPVSPASQKMVIQGPSSPQGEAMVTDVLEDQKEGRSTNKENPS
KALIERPSQNNIGIQTMECSLRVPETVSAATQTIKNVCEQGTSTVDQNFGKQDATVQTER
GSGEKPVSAPGDDTESLHSQGEEEFDMPQPPHGHVLHRHMRTIREVRTLVTRVITDVYYV
DGTEVERKVTEETEEPIVECQECETEVSPSQTGGSSGDLGDISSFSSKASSLHRTSSGTS
LSAMHSSGSSGKGAGPLRGKTSGTEPADFALPSSRGGPGKLSPRKGVSQTGTPVCEEDGD
AGLGIRQGGKAPVTPRGRGRRGRPPSRTTGTRETAVPGPLGIEDISPNLSPDDKSFSRVV
PRVPDSTRRTDVGAGALRRSDSPEIPFQAAAGPSDGLDASSPGNSFVGLRVVAKWSSNGY
FYSGKITRDVGAGKYKLLFDDGYECDVLGKDILLCDPIPLDTEVTALSEDEYFSAGVVKG
HRKESGELYYSIEKEGQRKWYKRMAVILSLEQGNRLREQYGLGPYEAVTPLTKAADISLD
NLVEGKRKRRSNVSSPATPTASSSSSTTPTRKITESPRASMGVLSGKRKLITSEEERSPA
KRGRKSATVKPGAVGAGEFVSPCESGDNTGEPSALEEQRGPLPLNKTLFLGYAFLLTMAT
TSDKLASRSKLPDGPTGSSEEEEEFLEIPPFNKQYTESQLRAGAGYILEDFNEAQCNTAY
QCLLIADQHCRTRKYFLCLASGIPCVSHVWVHDSCHANQLQNYRNYLLPAGYSLEEQRIL
DWQPRENPFQNLKVLLVSDQQQNFLELWSEILMTGGAASVKQHHSSAHNKDIALGVFDVV
VTDPSCPASVLKCAEALQLPVVSQEWVIQCLIVGERIGFKQHPKYKHDYVSH
|
|
|
|---|
| BDBM50496593 |
|---|
| n/a |
|---|
| Name | BDBM50496593 |
|---|
| Synonyms: | CHEMBL3134131 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H33N3 |
|---|
| Mol. Mass. | 327.5068 |
|---|
| SMILES | C(Cc1ccc(cc1)N1CCC(CC1)N1CCCC1)N1CCCC1 |
|---|
| Structure | 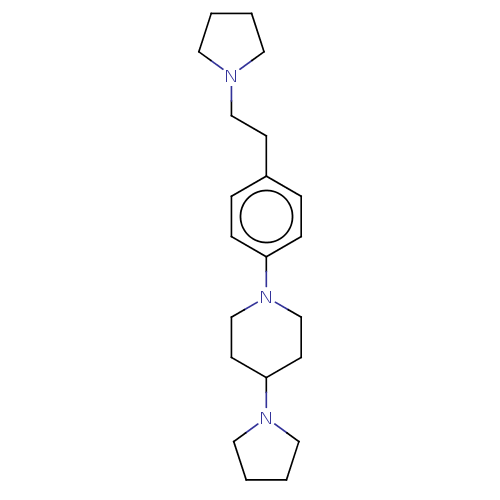
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Camerino, MA; Zhong, N; Dong, A; Dickson, BM; James, LI; Baughman, BM; Norris, JL; Kireev, DB; Janzen, WP; Arrowsmith, CH; Frye, SV The structure-activity relationships of L3MBTL3 inhibitors: flexibility of the dimer interface. Medchemcomm4:1501-1507 (2013) [PubMed] Article
Camerino, MA; Zhong, N; Dong, A; Dickson, BM; James, LI; Baughman, BM; Norris, JL; Kireev, DB; Janzen, WP; Arrowsmith, CH; Frye, SV The structure-activity relationships of L3MBTL3 inhibitors: flexibility of the dimer interface. Medchemcomm4:1501-1507 (2013) [PubMed] Article