| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM50046356 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_213037 (CHEMBL816975) |
|---|
| IC50 | 19±n/a nM |
|---|
| Citation |  Shuman, RT; Rothenberger, RB; Campbell, CS; Smith, GF; Gifford-Moore, DS; Gesellchen, PD Highly selective tripeptide thrombin inhibitors. J Med Chem36:314-9 (1993) [PubMed] Shuman, RT; Rothenberger, RB; Campbell, CS; Smith, GF; Gifford-Moore, DS; Gesellchen, PD Highly selective tripeptide thrombin inhibitors. J Med Chem36:314-9 (1993) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Alpha-trypsin chain 1 | Alpha-trypsin chain 2 | Beta-trypsin | Cationic trypsinogen | PRSS1 | Serine protease 1 | TRP1 | TRY1 | TRY1_HUMAN | TRYP1 | Thrombin & trypsin | Trypsin | Trypsin I | Trypsin-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 26557.80 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P07477 |
|---|
| Residue: | 247 |
|---|
| Sequence: | MNPLLILTFVAAALAAPFDDDDKIVGGYNCEENSVPYQVSLNSGYHFCGGSLINEQWVVS
AGHCYKSRIQVRLGEHNIEVLEGNEQFINAAKIIRHPQYDRKTLNNDIMLIKLSSRAVIN
ARVSTISLPTAPPATGTKCLISGWGNTASSGADYPDELQCLDAPVLSQAKCEASYPGKIT
SNMFCVGFLEGGKDSCQGDSGGPVVCNGQLQGVVSWGDGCAQKNKPGVYTKVYNYVKWIK
NTIAANS
|
|
|
|---|
| BDBM50046356 |
|---|
| n/a |
|---|
| Name | BDBM50046356 |
|---|
| Synonyms: | 1-(2-Amino-2-phenyl-acetyl)-pyrrolidine-2-carboxylic acid (1-formyl-4-guanidino-butyl)-amide | CHEMBL104305 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H28N6O3 |
|---|
| Mol. Mass. | 388.464 |
|---|
| SMILES | [#7]-[#6@@H](-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@H]-1-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6]=O)-c1ccccc1 |
|---|
| Structure | 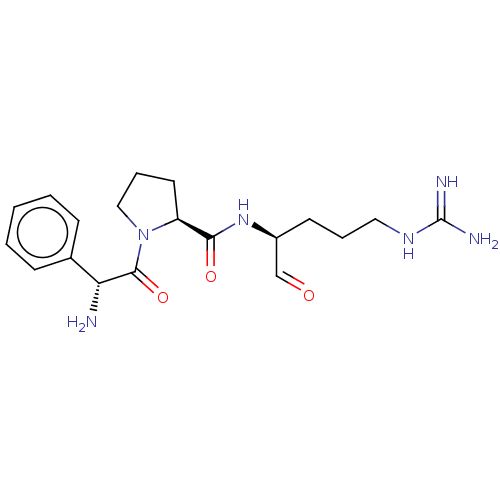
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Shuman, RT; Rothenberger, RB; Campbell, CS; Smith, GF; Gifford-Moore, DS; Gesellchen, PD Highly selective tripeptide thrombin inhibitors. J Med Chem36:314-9 (1993) [PubMed]
Shuman, RT; Rothenberger, RB; Campbell, CS; Smith, GF; Gifford-Moore, DS; Gesellchen, PD Highly selective tripeptide thrombin inhibitors. J Med Chem36:314-9 (1993) [PubMed]