

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Reverse transcriptase | ||
| Ligand | BDBM50532843 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1924015 (CHEMBL4426971) | ||
| IC50 | 580±n/a nM | ||
| Citation |  Stanton, RA; Lu, X; Detorio, M; Montero, C; Hammond, ET; Ehteshami, M; Domaoal, RA; Nettles, JH; Feraud, M; Schinazi, RF Discovery, characterization, and lead optimization of 7-azaindole non-nucleoside HIV-1 reverse transcriptase inhibitors. Bioorg Med Chem Lett26:4101-5 (2016) [PubMed] Article Stanton, RA; Lu, X; Detorio, M; Montero, C; Hammond, ET; Ehteshami, M; Domaoal, RA; Nettles, JH; Feraud, M; Schinazi, RF Discovery, characterization, and lead optimization of 7-azaindole non-nucleoside HIV-1 reverse transcriptase inhibitors. Bioorg Med Chem Lett26:4101-5 (2016) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Reverse transcriptase | |||
| Name: | Reverse transcriptase | ||
| Synonyms: | n/a | ||
| Type: | Protein | ||
| Mol. Mass.: | 29598.37 | ||
| Organism: | Human immunodeficiency virus 1 | ||
| Description: | Q9WKE8 | ||
| Residue: | 254 | ||
| Sequence: |
| ||
| BDBM50532843 | |||
| n/a | |||
| Name | BDBM50532843 | ||
| Synonyms: | CHEMBL4545462 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H17F4N3O | ||
| Mol. Mass. | 463.4263 | ||
| SMILES | CCCCn1cc(C#N)c2c(c3C(=O)c4ccccc4-c3nc12)-c1ccc(cc1F)C(F)(F)F |(40.53,-30.12,;39.2,-30.9,;39.2,-32.44,;37.86,-33.22,;37.87,-34.76,;39.12,-35.67,;38.64,-37.14,;39.55,-38.38,;40.45,-39.63,;37.1,-37.14,;36.07,-38.27,;34.57,-37.96,;33.33,-38.87,;33.34,-40.41,;32.08,-37.97,;30.59,-38.29,;29.55,-37.16,;30.02,-35.69,;31.52,-35.37,;32.55,-36.5,;34.09,-36.5,;35.12,-35.35,;36.62,-35.67,;36.55,-39.74,;38.05,-40.05,;38.54,-41.51,;37.51,-42.67,;36,-42.35,;35.52,-40.88,;33.97,-40.88,;37.99,-44.13,;36.96,-45.28,;39.5,-44.45,;38.38,-45.62,)| | ||
| Structure | 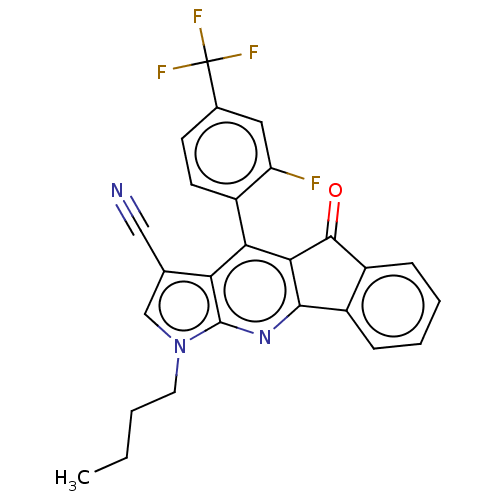
| ||