| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM28701 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2050198 (CHEMBL4704897) |
|---|
| EC50 | 31700±n/a nM |
|---|
| Citation |  Peiretti, F; Montanari, R; Capelli, D; Bonardo, B; Colson, C; Amri, EZ; Grimaldi, M; Balaguer, P; Ito, K; Roeder, RG; Pochetti, G; Brunel, JM A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor ? Binding Properties and Biological Activities. J Med Chem63:13124-13139 (2020) [PubMed] Article Peiretti, F; Montanari, R; Capelli, D; Bonardo, B; Colson, C; Amri, EZ; Grimaldi, M; Balaguer, P; Ito, K; Roeder, RG; Pochetti, G; Brunel, JM A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor ? Binding Properties and Biological Activities. J Med Chem63:13124-13139 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM28701 |
|---|
| n/a |
|---|
| Name | BDBM28701 |
|---|
| Synonyms: | 2-(4-{2-[(4-chlorophenyl)formamido]ethyl}phenoxy)-2-methylpropanoic acid | Azufibrat | Bezafibrate | CHEMBL264374 | Cedur | Sklerofibrat |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H20ClNO4 |
|---|
| Mol. Mass. | 361.819 |
|---|
| SMILES | CC(C)(Oc1ccc(CCNC(=O)c2ccc(Cl)cc2)cc1)C(O)=O |
|---|
| Structure | 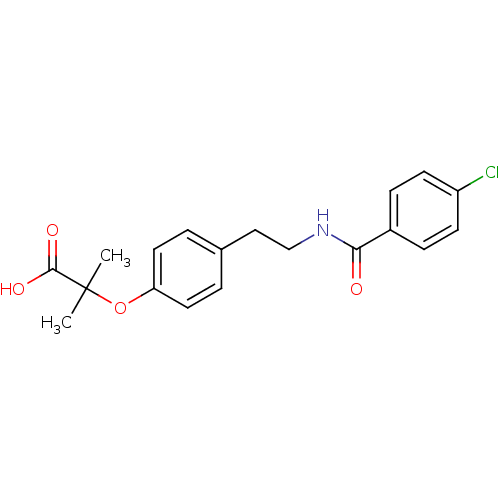
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Peiretti, F; Montanari, R; Capelli, D; Bonardo, B; Colson, C; Amri, EZ; Grimaldi, M; Balaguer, P; Ito, K; Roeder, RG; Pochetti, G; Brunel, JM A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor ? Binding Properties and Biological Activities. J Med Chem63:13124-13139 (2020) [PubMed] Article
Peiretti, F; Montanari, R; Capelli, D; Bonardo, B; Colson, C; Amri, EZ; Grimaldi, M; Balaguer, P; Ito, K; Roeder, RG; Pochetti, G; Brunel, JM A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor ? Binding Properties and Biological Activities. J Med Chem63:13124-13139 (2020) [PubMed] Article